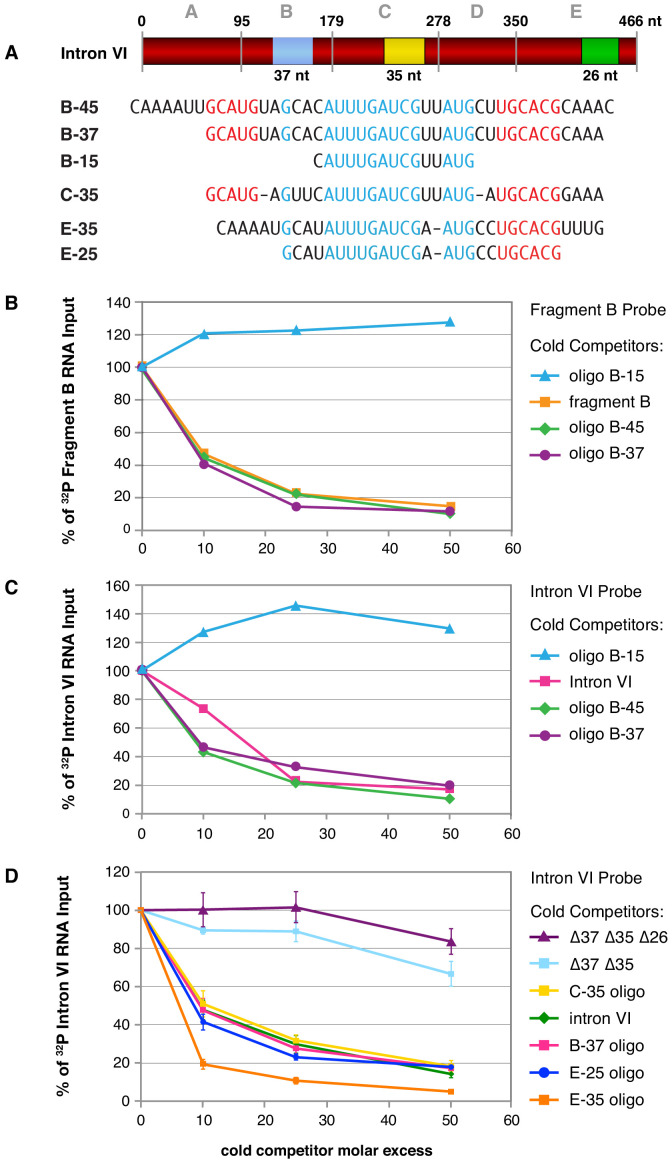
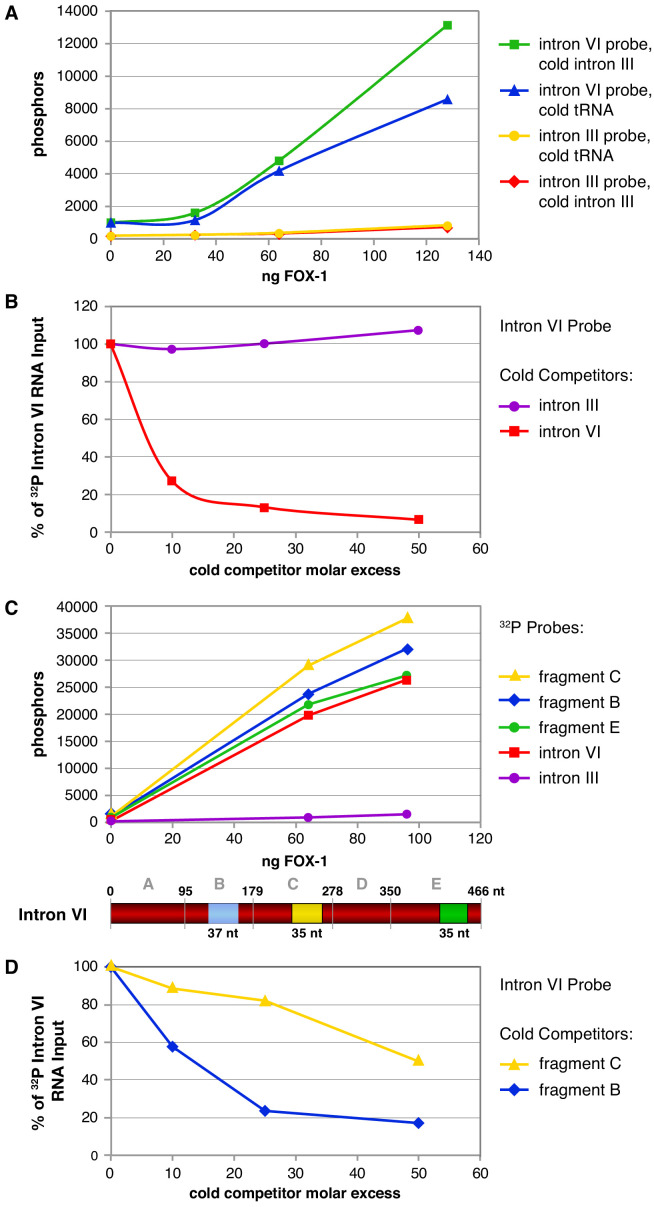
Figure 5. Purified FOX-1 protein binds in vitro to multiple sites in intron VI using motifs GCAUG and GCACG.
(A) The diagram of intron VI shows the intron VI fragments (A–E) and smaller regions (RNA oligonucleotides B-45 to E-25) tested for direct FOX-1 binding in vitro. Only RNAs from fragments B, C, and E bind to purified FOX-1 (Figure 5—figure supplement 1C). Motifs GCAUG and GCACG (red) and sequences common to all three fragments (light blue) are shown below the diagram. Blue, yellow, and green rectangles indicate the locations of sequences B-37, C-35, and E-25, respectively, within FOX-1 binding fragments of intron VI. The RNA oligonucleotides listed were used in competition experiments with 32P-labeled fragment B (panel B) and 32P-labeled intron VI (panels C and D). (B) Small RNA oligonucleotides corresponding to sequences within fragment B compete for FOX-1 binding in vitro. Graphs show cross-linking competition experiments in which binding of FOX-1 (32 ng) to 32P-labeled fragment B RNA was challenged with an increasing molar excess of either cold fragment B RNA or small RNA oligonucleotides to sequences in fragment B that are also found in the other FOX-1 binding regions in fragments C and E. Binding is expressed as the percent of 32P fragment B bound by FOX-1 without any competitor RNA. (C) RNA oligonucleotides compete for FOX-1 binding to intron VI. The cross-linking competition experiments are similar to those in panel (A), except the probe is 32P-labeled full-length intron VI RNA. Binding is expressed as the percent of 32P intron VI bound by FOX-1 without any competitor RNA. The finding that the B-15 oligonucleotide fails to compete with either fragment B probe or intron VI probe, while the B-45 and B-37 oligonucleotides compete well, indicates that GCAUG, GCACG, or both are utilized for FOX-1 binding. (D) FOX-1 binds to multiple sites within intron VI using both GCAUG and GCACG. Graphs show results of cross-linking competition experiments in which binding of FOX-1 (32 ng) to 32P-labeled intron VI RNA was challenged with an increasing molar excess of several cold RNAs, as indicated. Binding is expressed as the percent of 32P intron VI RNA bound by FOX-1 without any competitor RNA. Cold intron VI RNA carrying deletions of the common sequences in B (Δ37) and C (Δ35) competed very poorly with intron VI probe for FOX-1 binding, and cold intron VI with deletions in all three common regions [B (Δ37), C (Δ35), and E (Δ26)] competed even less efficiently, demonstrating the critical role of these sequences in FOX-1 binding. In contrast, RNA oligonucleotides (C-35, B-37, E-35, and E-25) of sequences in fragments B, C, and E competed very effectively with intron VI for binding to FOX-1, further supporting the conclusion that FOX-1 binds to multiple sites in intron VI. The 25 nt RNA oligonucleotide in fragment E contains only the motif GCACG, but not GCAUG, indicating that GCACG promotes robust FOX-1 binding. The deletion in E (Δ26) is one nucleotide longer than the E-25 oligonucleotide, including deletion of a 3' U. Error bars, SEM.