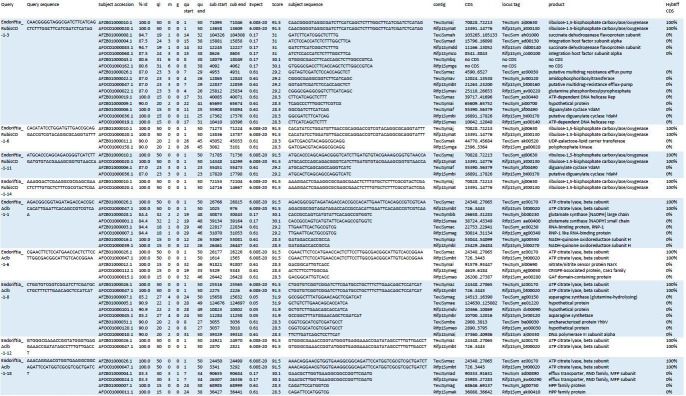
Author response image 1. Blast results of HCR-FISH probes and in silico analysis of their hybridization behaviour.
Four RubisCO-specific probes and five AclB probes were searched against the NCBI whole-genome shotgun (WGS) sequences NZ_AFOC00000000.1 and NZ_AFZB00000000.1, containing the complete genomes of the Riftia pachyptila symbiont and the Tevnia jerochonana symbiont, respectively (both symbionts belong to the same 16S rRNA phylotype; Gardebrecht et al ., 2012). At low stringency settings (BLASTn optimized for “somewhat similar sequences”, seed length: 7, expectation threshold: 1), most probe sequences (queries) had matches (subjects) not only in their respective target genes, but also produced off-target matches of lower % identity (%Id) and with higher numbers of gaps (g) and mismatches (m) in the alignment. All subject sequences were subjected to in silico analyses using the online tool mathFISH at simulated hybridization conditions matching those in our experiments (25% formamide, 45°C, 2 nM probe concentration). As a result, 100% hybridization efficiency was predicted for probe binding to the respective target sequences, whereas binding to off-target matches was calculated with 0% hybridization efficiency. ql: query length; qu start: start of alignment in query; qu end: end of alignment in query; contig numbers beginning with “TevJSym” refer to the the Tevnia symbiont genome assembly, contig numbers beginning with “Rifp1Sym” refer to the Riftia symbiont genome assembly; CDS: complete coding sequence from start to end; product: protein encoded by coding sequence; HybEff CDS: hybridization efficiency of the probe/target pair (probe = query sequence; target = complete CDS of subject).