Figure 1. LUCS sampling strategy to create de novo designed protein fold families with tunable geometries.
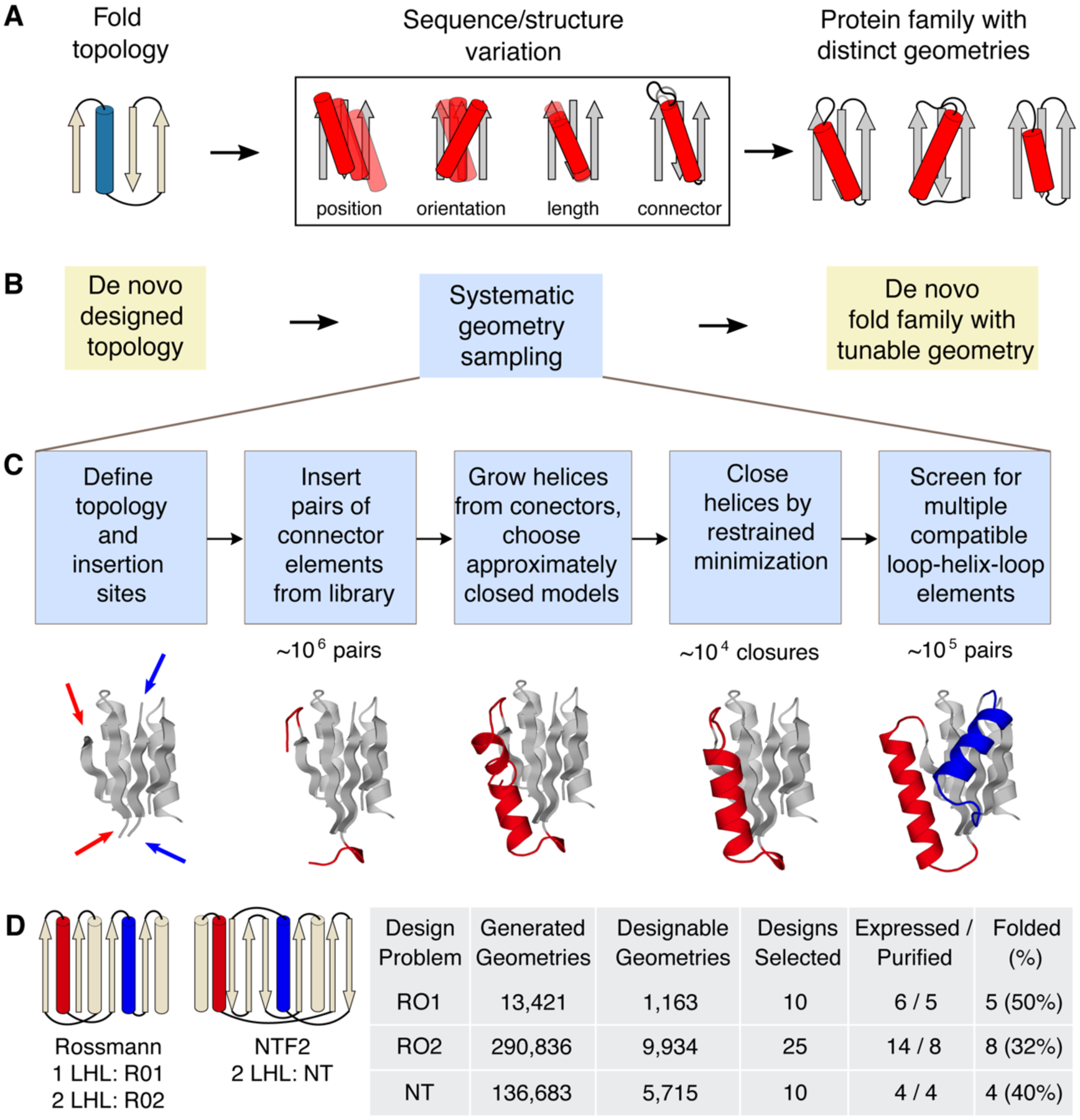
A. In nature, protein fold topologies (left) are diversified to create families of proteins with distinct geometries (right) optimized for function. Alpha-helices are shown as cylinders and beta-strands as arrows. The box shows schematic representations of common types of geometric variation. B. The LUCS computational design protocol seeks to mimic the ability of evolution to diversity protein geometries to generate de novo designed fold families. C. Schematic of the LUCS protocol for sampling LHL geometries. The reshaped LHL units are colored in red and blue. Typical numbers of models generated at major stages of the protocol are indicated. D. Designed fold families. Schematic shows fold topologies and design problems (Rossman fold with 1 or 2 reshaped LHL units, and NTF2 fold with 2 reshaped LHL units). Also shown are numbers for geometries generated by LUCS, designed models that passed quality filters, and experimentally characterized designs for three design problems. % folded indicates the fraction of experimentally tested designs that adopted folded structures.
