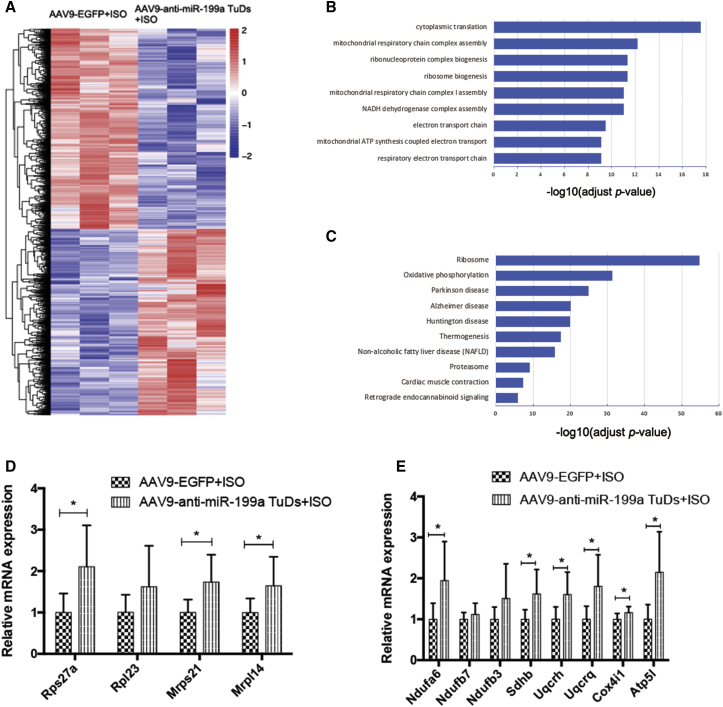
Figure 4.
RNA sequencing results of anti-miR-199a treatment in cardiac hypertrophy
(A) Heatmap of all genes with significantly altered expression between the cardiac hypertrophy group and the anti-miR-199a-treated group. Red and blue colors indicate upregulated or downregulated genes. n = 3 per group. (B) Gene Ontology (GO) enrichment analysis of all significantly upregulated genes. The y axis shows the top 10 enriched pathways. The x axis is the −log10 of the adjusted p value. (C) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of all significantly upregulated genes. The y axis shows the top enriched pathways. The x axis is the −log10 of the adjusted p value. (D) The expression of representative ribosome genes in the cardiac hypertrophy group and the anti-miR-199a-treated group. n = 8. (E) The expression of representative genes involved in oxidative phosphorylation in the cardiac hypertrophy group and the anti-miR-199a-treated group. n = 8 per group. ∗p < 0.05, ∗∗p < 0.01.