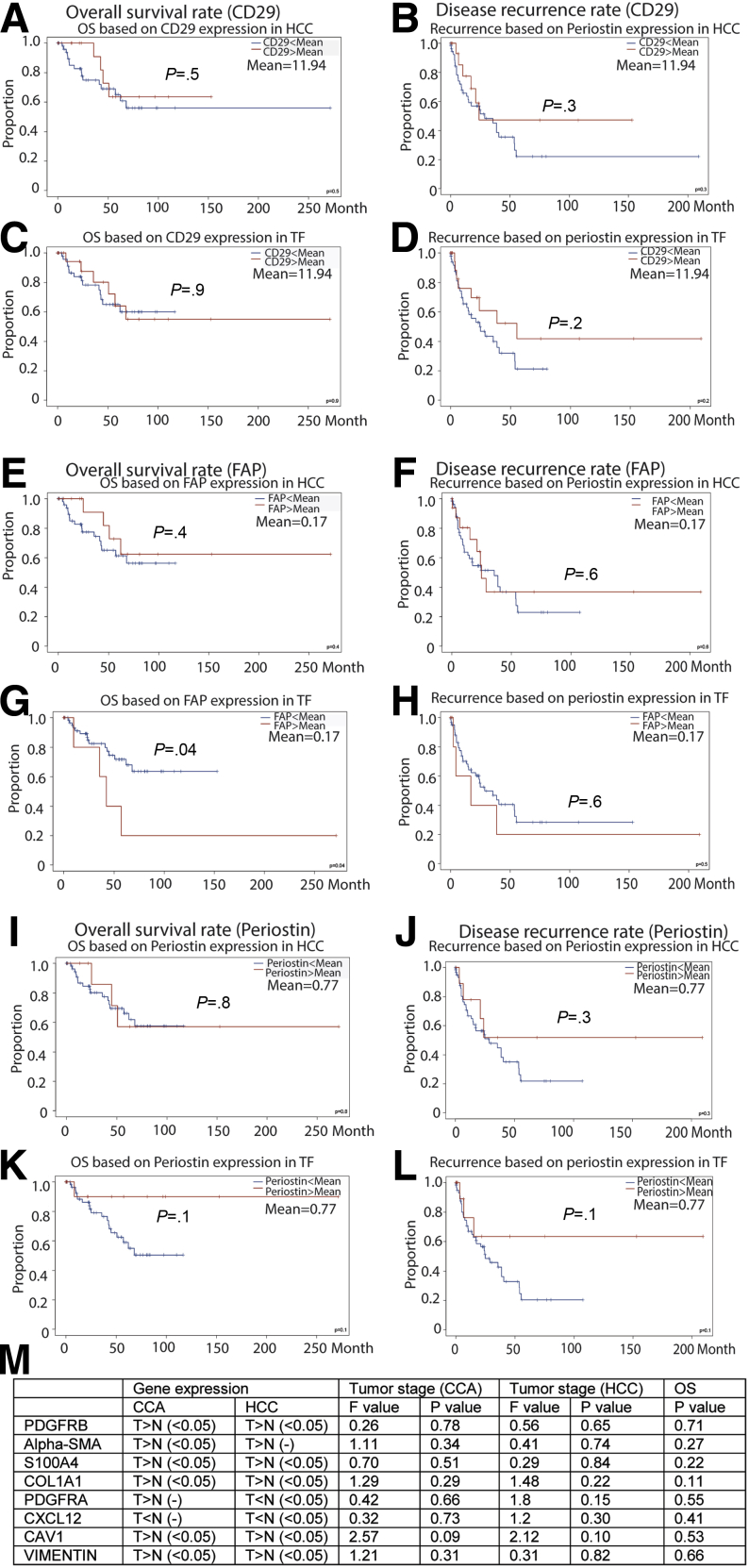
Figure 2.
Survival and recurrence analysis based on the gene expression of CD29, FAP, and periostin in our HCC cohort and bioinformatics analysis of other CAF markers in the GEPIA online database. (A–L) Overall survival and disease-free rate based on the gene expression of CD29, FAP, and periostin in tumor tissue or tumor-free liver tissue of our HCC patients (Kaplan–Meier analysis, N = 75). (M) Bioinformatics analysis of PDGFRB, α-SMA, S100A4, COL1A1, PDGFRA, CXCL12, CAV1, and vimentin in the GEPIA database. The gene expression of these markers in tumor and normal liver tissue (CCA: n = 9 for normal liver tissue; n = 36 for tumor tissue; HCC: n = 160 for normal liver tissue; n = 369 for tumor tissue) was assessed by 1-way analysis of variance. Gene expression of these markers in different stages of liver tumors (CCA: n = 36; HCC: n = 369) was assessed by 1-way analysis of variance. The differences in survival related to CAF markers PDGFRB, α-SMA, FSP1, COL1A1, PDGFRA, CXCL12, CAV1, and vimentin messenger RNA expression were compared in each group involving all patients (Log-rank test, n = 36 for CCA, n = 364 for HCC). (-) Without a statistically significant difference. CAV1, caveolin 1; COL1A1, collagen type I α 1; CXCL12, C-X-C motif chemokine ligand 12; FSP1, fibroblast-specific protein 1; N, normal liver tissue; OS, overall survival; PDGFRB, platelet-derived growth factor receptor β; T, tumor tissue; TF, tumor free.