Figure 1.
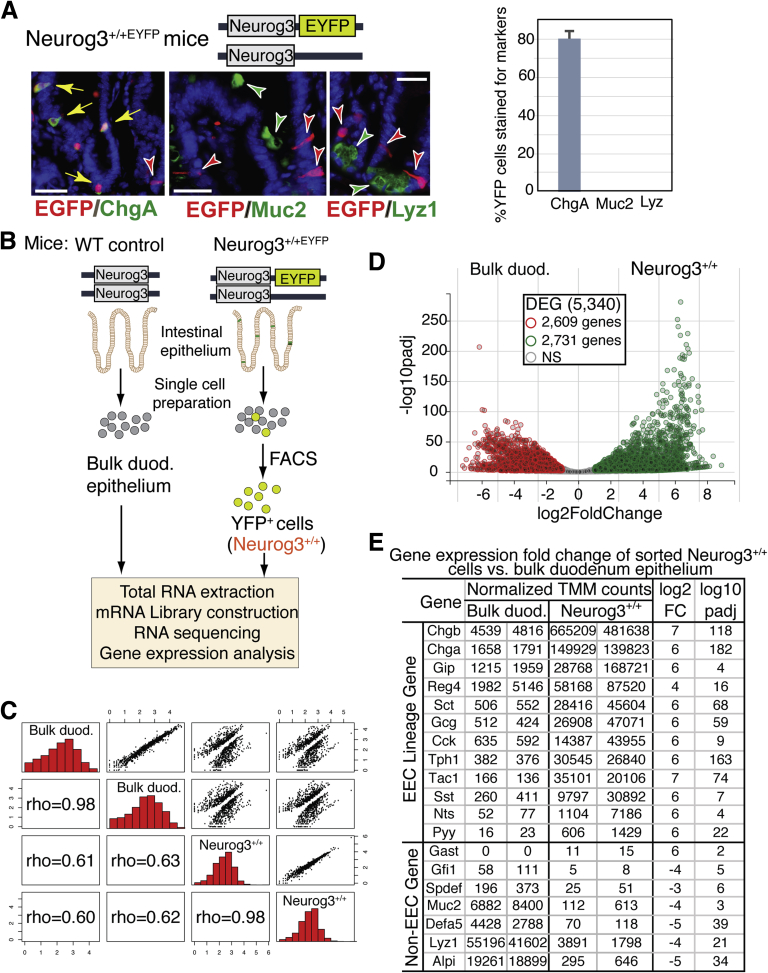
EYFP expression is restricted to EECs in the small intestine of Neurog3+/+EYFP(Neurog3+/+) reporter mouse. (A) Top: Schematic diagram of Neurog3 and EYFP at the Neurog3 locus in Neurog3+/+EYFP mouse. Bottom: Adult mouse intestinal sections from Neurog3+/+ mice were co-stained with antibodies against GFP (in red) and against ChgA, Muc2, or lysozyme, markers (in green) for enteroendocrine, goblet, and Paneth cells, respectively. Yellow arrows indicate co-expression, and 4′,6-diamidino-2-phenylindole (in blue) counterstained for nuclei. Scale bars: 50 um. Percentage of GFP cells stained for ChgA, Muc2, or Lyz was summarized in the bar graph (right) (N = 3). P < .00001 as indicated. The P value was calculated by 1-tailed Student t test. (B) Experimental flow chart of cell isolation and RNA-seq procedure. (C) All2all scatter plot of all transcripts detected after batch correction: an overview of similarities and variance between samples sequenced. Each Neurog3+/+ library was made from RNAs isolated and sorted duodenum cells from 2 Neurog3+/+EYFP mice. Each bulk duodenum (Bulk duod.) library was made from RNAs from 1 wild-type (WT) control mouse duodenum epithelial cells. Two independent replicates were performed for each genotype. The upper side of the plot shows a log10 normalized scatter plot. The bottom side of the plot is the Pearson correlations of each paired sample. Diagonal shows the histogram of normalized read counts in log10 scale. (D) Volcano plots of normalized TMM counts comparing the transcriptomes of sorted Neurog3+/+ cells and control intestinal epithelial cells (Unsorted). A total of 5340 transcripts were found to be expressed differentially (|Fc| ≥2), with 2609 genes enriched in the control cell population and 2731 genes enriched in the Neurog3+/+ cell population. (E) Selected transcript levels of EEC-specific hormones and marker genes expressed in Neurog3+/+ cells vs control unsorted intestinal cells. Normalized gene expression values (TMM) were adjusted using edgeR testing methods that account for library depth, RNA composition, and gene length among samples. Fc, fold change.