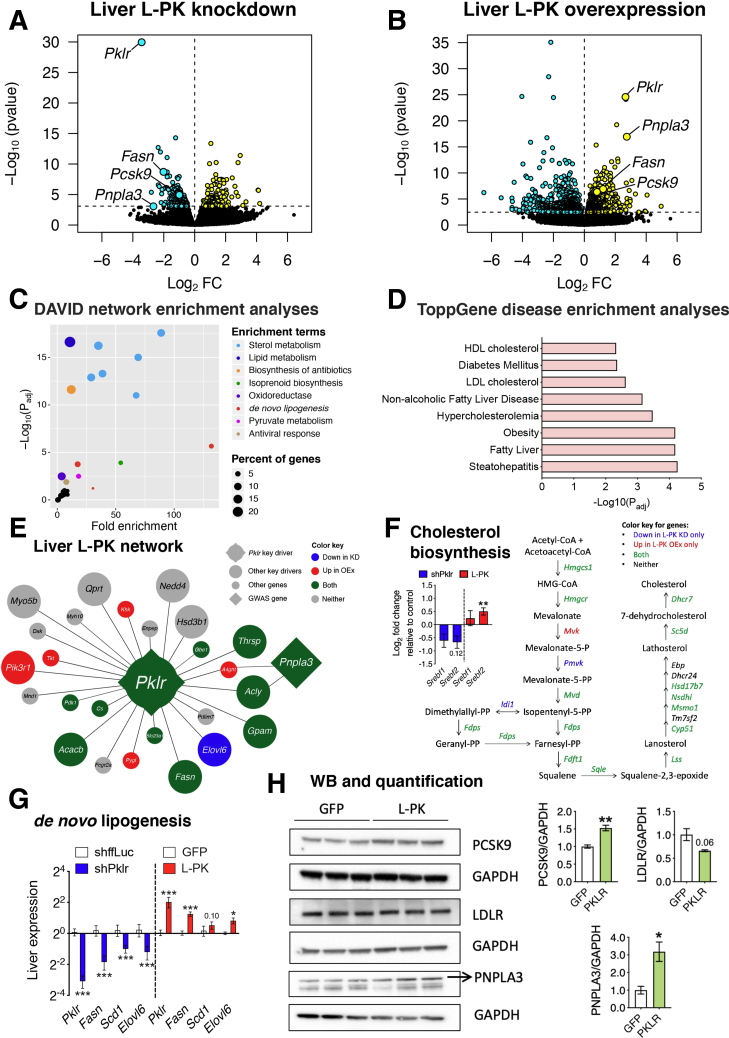
Figure 8.
L-PK alters liver steatosis via de novo lipogenesis and PNPLA3 and plasma cholesterol levels via PCSK9. Global genome-wide transcriptomics of livers extracted from L-PK (A) KD or (B) OEx mice. Turquoise and yellow represent down-regulated and up-regulated genes, respectively. (C) DAVID pathway and (D) ToppGene disease enrichment analyses of 124 common DEGs (Supplementary Table 1) between L-PK KD and OEx mice. Overlay of DEGs on (E) L-PK key driver network from our previous study9 or (F) cholesterol biosynthesis pathway. Blue represents genes going down in KD mice, red represents genes going up in OEx mice, and green represents changing in both mice (down in KD, up in OEx). Inset shows log2 fold change of Srebf1 and Srebf2 in L-PK KD and OEx mice. Follow-up (G) qPCR analyses of liver DNL genes such as Pklr, Fasn, Scd1, and Elovl6 and (H) immunoblot analyses of liver proteins such as PCSK9, LDLR, and PNPLA3 in L-PK OEx mice. Data are presented as mean ± SEM (n = 5 livers for RNA and 3 for protein analyses per group). ∗P < .05; ∗∗P < 0.1; ∗∗∗P < .001 by t test. KD, knockdown; LDL, low-density lipoprotein; OEx, overexpressing.