Figure EV1. Pervasive and dynamic alternative splicing occurs during human hematopoietic differentiation induced from hESCs.
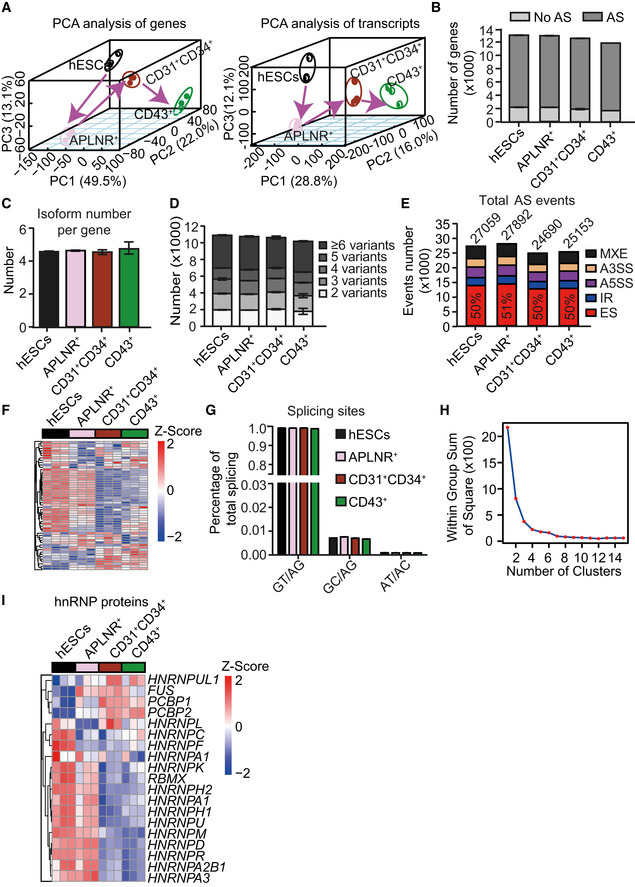
- Principal component analysis (PCA) of genes (left) and transcripts (right) in highly purified cells at various differentiation stages, including ESCs, APLNR+ cells, CD31+CD34+ cells, and CD43+ cells during hematopoietic differentiation induction from hESCs.
- Widespread occurrence (> 85% of expressed genes) of alternative splicing in expressed genes (FPKM> 1 in at least one differentiation stage) at distinct differentiation stages during hematopoietic development.
- Average number of isoforms per gene at each differentiation stage.
- Analysis of isoform variants within each expressing gene at distinct differentiation stages.
- Number and frequency of five major splicing events at distinct differentiation stages, including mutually exclusive exon (MXE), alternative 5′ splicing (A5SS), alternative 3′ splicing (A3SS), intron retention (IR), and exon skipping (ES). The cutoff of splicing event of an expressed gene is 0.05 < PSI < 0.95.
- Heatmap showing the genes within the minor spliceosomal machinery (n = 52) at the indicated stages. The heatmap was row normalized. The heatmap was scaled with Z‐Score using the log2(FPKM) expression of indicated components within the minor spliceosomal machinery.
- Frequency of splicing site at different differentiation stages.
- The elbow plot assessing the potential clusters of the dynamic splicing factors expression in the heatmap of Fig 1 (C).
- Heatmap showing the genes in the hnRNP family at the indicated stages. The heatmap was row normalized. The heatmap was scaled with Z‐Score using the log2(FPKM) expression of indicated genes..
Data information: Results given are mean ± SD.For all panels n = 3 technical replicates.
