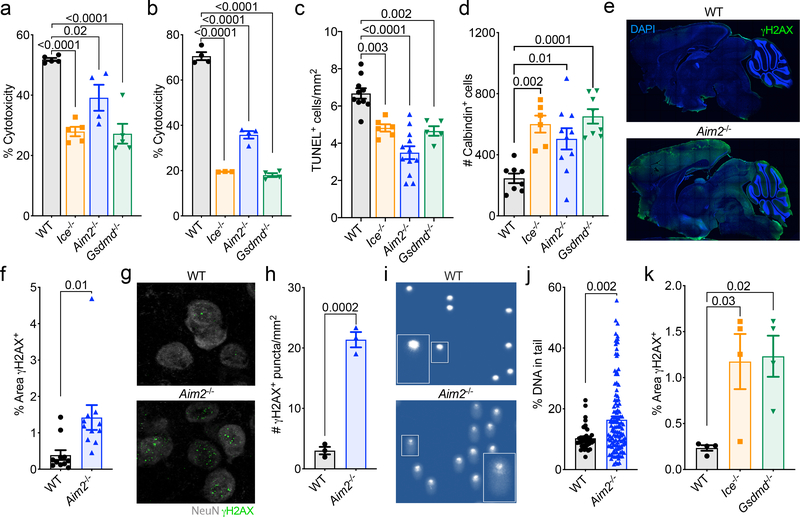
Figure 3. Activation of the AIM2 inflammasome in response to DNA damage coordinates CNS cell death and limits the accumulation of DNA damage in the brain.
(a-b) Postnatal day 0 (p0) mixed neural cultures were primed with LPS for 4 hrs followed by overnight treatment with either (a) 40 Gy ionizing radiation (WT n=5, Ice−/− n=5, Aim2−/− n=4, and Gsdmd−/− n=5) or (b) 100 μM etoposide (WT n=4, Ice−/− n=3, Aim2−/− n=4, and Gsdmd−/− n=4). Cell death was measured by LDH release. Representative data from 3 independent experiments. (c) Quantification of cerebellar TUNEL staining in p5 mice (WT n=10, Ice−/− n=6, Aim2−/− n=12, and Gsdmd−/− n=5 mice; from 3 independent experiments). (d) Enumeration of cerebellar calbindin+ Purkinje neurons in adult (8–12 weeks old) mice (WT n=8, Ice−/− n=6, Aim2−/− n=10, and Gsdmd−/− n=8 mice; from 3 independent experiments). (e-f) Adult brains were evaluated for levels of DNA damage (γH2AX, green). (e) Representative images from 3 independent experiments with similar results. (f) Quantification of γH2AX staining in adult mice (WT n=11, Aim2−/− n=11; from 3 independent experiments). (g-h) Adult brains were evaluated for γH2AX staining in NeuN-expressing neurons in the amygdala. (g) Representative images from 2 independent experiments with similar results. (h) Enumeration of γH2AX puncta in the amygdala (WT n=3, Aim2−/− n=3; representative data from 2 independent experiments). (i-j) DNA damage was evaluated in the cortex of 10-week-old WT and Aim2−/− mice by comet assay. (i) Representative images of single cell electrophoresis gels from 3 independent experiments with similar results. (j) Quantification of percent DNA in tail (WT n=38, Aim2−/− n=120; from 3 independent experiments). (k) Quantification of γH2AX staining in sagittal brain sections from WT n=4, Ice−/− n=4, and Gsdmd−/− n=4 mice; from 2 independent experiments. Error bars depict mean ± s.e.m. Statistics calculated by (a-d,k) one-way ANOVA with Tukey’s post hoc tests and (f,h,j) unpaired two-tailed Student’s t-test. (a-b,j) n values refer to biological replicates from representative experiments. (c-d,f,h,k) n values refer to the number of mice used.