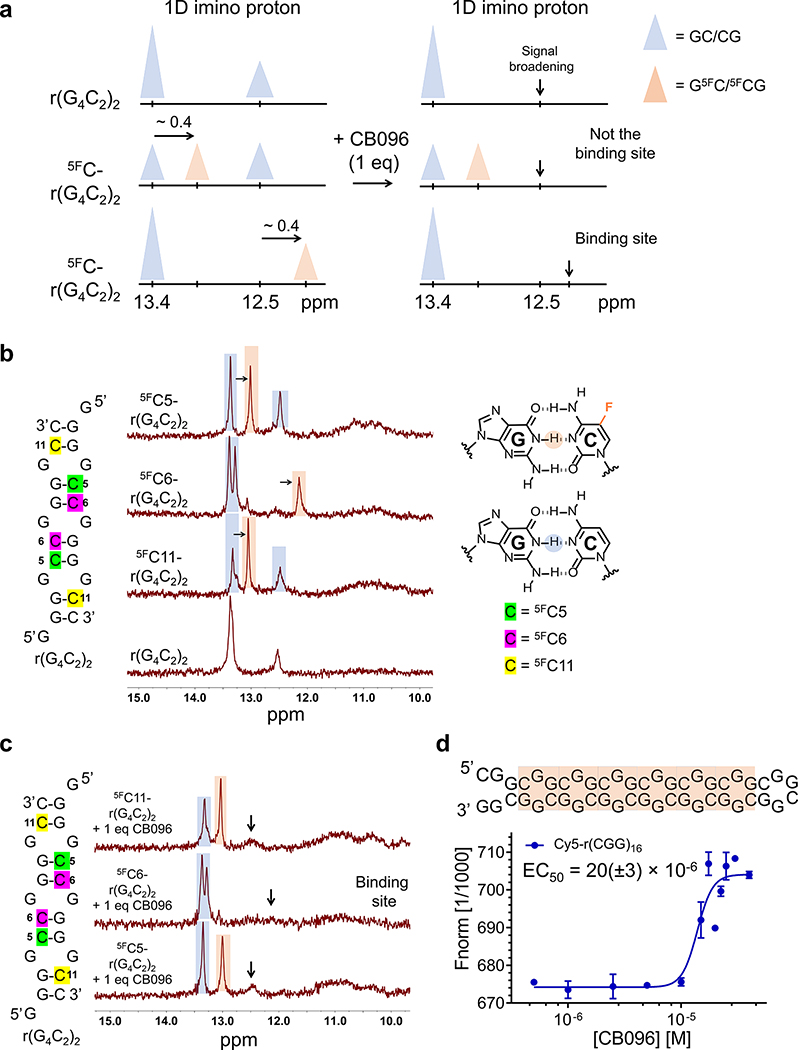
Figure 3.
CB096 affects the dynamics of 5′CGG/3′GGC, but not 5′GGC/3′CGG within r(G4C2)2. (a) Graphical representation of the strategy to assign the imino proton signals emanating from guanine (G) residues in base pairing interactions within r(G4C2)2. Briefly, replacing cytosine (C) by 5-fluorocytosine (5FC) shifts imino proton signals of G residues in GC base pairs upfield by ∼0.4 ppm (orange triangle), compared to unlabeled base pair peaks (blue triangle). (b) 1D imino proton spectra of 5FC labeled r(G4C2)2 duplex constructs. Blue color highlights the imino proton peak corresponding to the GC base pairs while those labeled in orange are from 5FC-labeled pairs. The 1D NMR spectra are representative of two independent experiments. (c) 1D imino proton spectra of 5FC labeled r(G4C2)2 duplex constructs in complex with CB096 (1 equiv). Black arrows indicate the imino proton peaks that broadened out upon CB096 treatment. The data indicate that CB096 binds 5′CGG/3′GGC and increases the dynamics of the base pairs (likely do not form or form transiently). The 1D NMR spectra are representative of two independent experiments. (d) CB096 binds to 5′CGG/3′GGC within Cy5-r(CGG)16 (orange squares) with an EC50 of 20(±3) μM, as measured by MST. The EC50 value was determined by plotting the changes in normalized fluorescence upon dose-dependent addition of CB096. The data points are reported as the mean values ± SD and are representative of two independent experiments performed in duplicate.