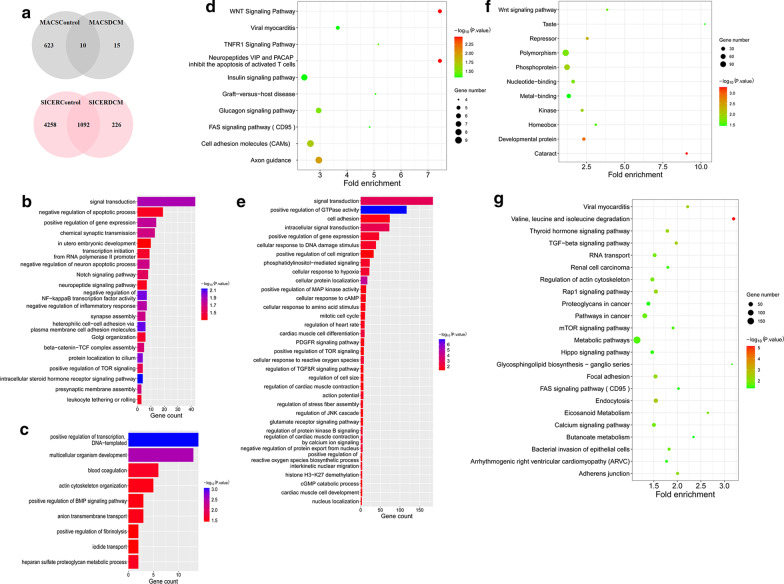
Fig. 2.
Identification of ChIP-seq differential binding genes, GO annotation, and pathway analysis. a Venn diagram indicating the overlap from MACS and SICER between control and DCM samples, respectively. b-d Biological process GO terms of differential target genes. b LMNA differential target genes in normal tissues. c Differential target genes of LAP2α-lamin A/C complexes in DCM. d Differential target genes of LAP2α-lamin A/C complexes in control samples. e–g Pathway analysis of differential target genes. The color of the circle represents the p value, and the size of the circle refers to the number of genes enriched in the same entry. e LMNA differential target genes in normal tissues. f Differential target genes of LAP2α-lamin A/C complexes in DCM. g Differential target genes of LAP2α-lamin A/C complexes in control samples