Abstract
The ability of tumor cells to spread from their origin place and form secondary tumor foci is determined by the epithelial–mesenchymal transition process. In epithelial tumors such as prostate cancer (PCa), the loss of intercellular interactions can be observed as a change in expression of polarity proteins. Epithelial cells acquire ability to migrate, what leads to the formation of distal metastases. In recent years, the interest in miRNA molecules as potential future treatment options has increased. In tumor microenvironment, miRNAs have the ability to regulate signal transduction pathways, where they can act as suppressors or oncogenes. MiRNAs are secreted by cancer cells, and the changes in their expression levels are closely related to a cancer progression, including epithelial–mesenchymal transition. These molecules offer new diagnostic and therapeutic possibilities. Therapeutics which make use of synthesized RNA fragments and mimic or block miRNAs affected in PCa, may lead to inhibition of tumor progression and even disease re-emission. Based on appropriate qualification criteria, we conducted a selection process to identify scientific articles describing miRNAs and their relation to epithelial–mesenchymal transition in PCa patients. The studies were published in English on Pubmed, Scopus and the Web of Science before August 08, 2019. Hazard ratios (HRs) and 95% confidence intervals (CI) as well as total Gleason score were used to assess the concordance between miRNAs and presence of metastases. A total of 13 studies were included in our meta-analysis, representing 1608 PCa patients and 15 miRNA molecules. Our study clarifies a relationship between the clinicopathological features of PCa and the aberrant expression of several miRNA as well as the complex mechanism of miRNA molecules involvement in the induction and promotion of the metastatic mechanism in PCa.
Keywords: Epithelial–mesenchymal transition, EMT, Metastases, microRNA, miRNA, Prostate cancer, Meta-analysis
Introduction
Prostate cancer (PCa) is the second most common diagnosed cancer and is considered the fifth leading cause of cancer-related deaths among men worldwide [1]. According to American Cancer Society statistics, the PCa incidence rates between 2011 and 2015 amounted 109.2 per year and the PCa death rates between 2012 and 2016 reached 19.2, showing decrease tendency [2] with the peak incidence (237.5 per year) and death rates (39.2) in 1992 [3]. In general, PCa has outstanding survival rates [3]. Surveillance, Epidemiology, and End Results Program (SEER 18 2009–2015) data shows the relative 5-year survival rate of local and regional stage PCa is almost 100%, for all races [2, 3]. However, for late stage PCa, the relative 5-year survival rate is only 30.5% [2, 3] Moreover, it is estimated that about 30% of men with diagnosed PCa have bone metastases [4]. In the U.S., the estimations for PCa with bone metastases amount to 6.5% (47,607 PCa with bone metastases in 735,812 of all PCa cases) of all PCa cases in 2004 [5]. Since 2007, the low-risk PCa incidence rate has decreased, however, the metastatic PCa incidence rate has notable increased [6].
The metastatic PCa involves multistep processes, including invasion, migration and survival of migrating cells, also location and proliferation in distant sites [7]. A critical point in metastases formation is attributed to the epithelial–mesenchymal transition (EMT) process [8]. EMT is a unique process by which tumor epithelial cells undergo molecular reprogramming and cytoskeleton remodeling through their transition to mesenchymal cell phenotype, The cells lose their adhesion capability and acquire the ability to move and to invade other tissues [9]. It is well known that the EMT process constitutes the early stage of metastases [10]. More specifically, the EMT transition is characterized by E-cadherin suppression associated with loss of cell adhesion properties and acquisition of mesenchymal markers, such as N-cadherin, Vimentin, Fibronectin, which enable cell mobility [11].
In the last decade, microRNAs have been shown to play an important role in managing single or numerous complex steps of metastases leading to tumor outgrowth by deregulation of target genes expression. MicroRNAs (miRNAs) are approximately 18–22 nucleotides long, single-stranded and non-coding regulatory RNAs, habitually aberrant in PCa [12, 13]. In normal tissue, microRNAs have the ability to regulate many biological processes such as cell proliferation, cell cycle arrest, aging and apoptosis. A number of studies have confirmed their presence and participation in metastases. They are responsible for controlling the expression level of prometastatic genes through base-paring with messenger RNAs (mRNAs) at the post-transcriptional level and lead to enhancement or suppression of the metastatic process.
Single miRNA has the ability to target multiple mRNA genes simultaneously. Many previous studies have revealed that above 50% of miRNA genes are situated in cancer-associated genomic regions and they form a central nodal points due to cancer progression [14]. They constitute crucial modulators in human cellular signaling pathways and are engaged in controlling gene expression of their targets. The abnormal expression level of miRNAs is heavily related to disease progression [15]. Owing to changes in expression levels, miRNAs have been classified as oncogenic molecules and tumor suppressors. Oncomirs (Onc) promote cancerogenesis through damping the mRNAs encoding tumor suppressor proteins at translational level (miR-301a, miR-409-3p, miR-210-3p, miR-543, miR-409-3p, miR-410-3p in PCa). While, tumor suppressors (TS) miRNAs are responsible for repression of mRNAs encoding onco-proteins at translational level (miR-573, miR-802, miR-3622a, miR-466, miR-203, miR-132/212, miR-141-3p in PCa).
MicroRNAs show strong promise as molecular biomarkers and treatment opportunity due to several aspects. Firstly, they manifest tissue-specific expression patterns in malignancies and exhibit a highly stable form in formalin-fixed tissues [16], as well as in blood, plasma and other bodily fluids [17, 18]. Circulating microRNAs are resistant to endogenous RNases activity, therefore their expression level can be determined in bodily fluids. Secondly, miRNA expression levels are varied in individual cancers, so they can specify different types of cancer. Thirdly, there has been observed a clear dissimilarity of miRNA regulation between different kinds of cancer. Fourthly, the efficiency and accuracy of miRNAs oriented to the target cells, make them attractive molecules in miRNA-based therapy. Fifthly, various microRNA profiles could be used for distinguish pathological and normal tissue as well as accurately recognize cancerous subtypes [19].
To the best of our knowledge there is no meta-analysis which describes the prognostic significance of miRNAs-related to the EMT process in PCa. In this study, we revealed the importance of fifteen deregulated miRNAs that constitute therapeutic potential and could be utilized as measurable indicators of PCa metastatic potential. To investigate their significance in EMT transition, we performed a meta-analysis, based on patient data extracted from scientific articles, where studies were conducted on formalin-fixed paraffin-embedded or fresh/snap-frozen tissues.
Materials and methods
Search strategy and eligibility criteria
The selected publications were identified by using up-to-date electronic databases, including PUBMED, Scopus and Web of Science online databases. The following keywords were used for the search: miRNA, EMT and prostate cancer. The inclusion criteria were as follows: (1) type of microRNA (miRNA was related to EMT process); (2) the articles were published between 2010 and 2019 year; (3) the study was conducted on formalin-fixed paraffin embedded tissue samples (FFPE) or fresh/snap-frozen prostate cancer tissues; (4) the study contained the following clinicopathological descriptions: TNM stages, according to TNM Classification of Malignant Tumors system (TNM) as well as Gleason score (GS) which were grouped by both low and high level of microRNAs expression. Exclusion criteria were as follows: (1) review articles; (2) studies based on PC cell lines; (3) non-clinical samples; (4) lacked sufficient data for estimating Hazard Ratios (HRs) and their 95% confidence intervals (CIs); (5) duplicate articles; (6) non-English paper; (7) case reports, letters, commentaries, conference abstract. The publication list was last updated on August 08, 2019.
Quality assurance
Methodology quality evaluation of incorporated studies was performed by two reviewers, working independently with the Newcastle–Ottawa Scale (NOS) for case–control quality assessment [20]. The evaluation process was divided into three steps: selection with a maximum score of 4, comparability with a maximum score of 2 and exposure/outcome with a maximum score of 3. A total score of 6–9 points ensured the highest quality.
Data extraction
The data extraction process was provided irrespectively two times by one author (MP) and was supervised by the senior reviewers in case of discrepancies (MB, DG). We collected the following items extracted from all included publications: (1) basic details: the title, first author’s last name, year of publication, country/continent of origin; (2) general details of the study: the name of microRNAs, sample size and kind of sample, microRNA detection method, size of both the low and high group of microRNAs expression level; (3) clinicopathological details: the TNM Classification of Malignant Tumors and Gleason score.
Statistical analysis
All statistical analyses were completed with the PQStat package. P value < 0.05 was considered as statistically significant. Estimation of miRNAs expression in PCa patients and their prognostic effects was presented using forest plots. We used the fixed-effects model to calculate Relative Risks (RRs) and 95% CIs in all included studies. In general, a RR > 1 demonstrated a poor patient outcome in cases with higher miRNA expression. The Cochrane’s Q statistic and Inconsistency index (I2) test was used to assess of heterogeneities. An I2 ≥ 50% is considered as a high heterogeneity level. Funnel plots were used to visualize of publication bias.
Results
Study characteristics
Study selection process identified 13 articles, which were included in the meta-analysis. Our study worked with articles published between 2010 and 2019, focusing on data of following miRNA: miR-410-3p [21], miR-573 [22], miR-3622a [23], miR-301a [24], miR-466 [25], miR-503 [26], miR-203 [27], miR-210-3p [28], miR-802 [29], miR-543 [30], miR-132 and miR-212 [31], miR-409-3p and miR-409-5p [32] and miR-141-3p [33].
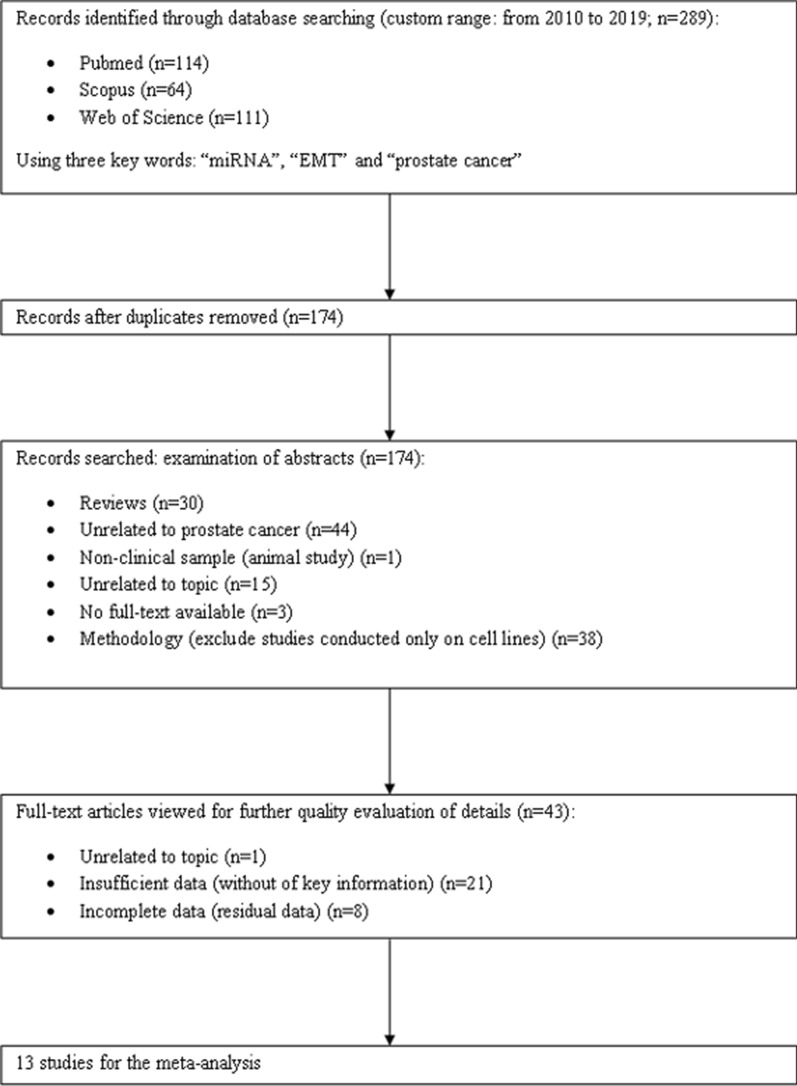
The total of 289 primary articles were searched in PubMed, Scopus and Web of Science databases. The flow chart diagram of the study selection process in our meta-analysis is presented below (Fig. 1). In the first step, we excluded 115 duplicated literatures. After examination of abstracts and full-text articles, we removed 161 publications and included 13 articles, finally. The main characteristics of involved studies was recapitulated in Table 1. The miRNAs in Table 1 were arranged in alphabetical order and some articles were divided into two parts because of more described miRNAs. The total group of the meta-analysis incorporated 1608 patients. The enrolled participants were divided into clinical and pathological stages, according to the TNM Classification of Malignant Tumors (TNM) standard. The participants of included studies mainly derived from China and the USA, but the dominant ethnicity was Chinese in approximately 60% of all studies. Detection of miRNAs deregulation was accomplished using real-time polymerase chain reaction (RT-PCR). The miRNAs fraction was isolated from fresh/snap-frozen or formalin-fixed paraffin embedded sections of tissue.
Fig. 1.
The flow diagram of meta-analysis
Table 1.
The main characteristic of included studies
| Author | Publication year | Country | miRNA type | General details of the study | ||
|---|---|---|---|---|---|---|
| Sample type | Sample size | Detection method | ||||
| Bucay | 2017 | USA | miR-3622a | FFPE | 138 | RT-PCR |
| Colden | 2017 | USA | miR-466 | FFPE | 96 | RT-PCR |
| Du | 2017 | China | miR-543 | Fresh | 42 | RT-PCR |
| Fu | 2016 | China | miR-132 | FFPE | 57 | RT-PCR |
| Fu | 2016 | China | miR-212 | FFPE | 57 | RT-PCR |
| Huang | 2017 | China | miR-141-3p | Snap-frozen | 141 | RT-PCR |
| Jiang | 2016 | China | miR-503 | FFPE | 82 | RT-PCR |
| Josson | 2017 | USA | miR-409-3p | Tissue microarray | 61 | RT-PCR |
| Josson | 2017 | USA | miR-409-5p | Tissue microarray | 61 | RT-PCR |
| Nam | 2016 | Canada | miR-301a | FFPE | 585 | RT-PCR |
| Ren | 2017 | China | miR-210-3p | Snap-frozen | 149 | RT-PCR |
| Saini | 2010 | USA | miR-203 | FFPE | 22 | RT-PCR |
| Wang | 2015 | China | miR-573 | FFPE | 80 | RT-PCR |
| Wang | 2017 | China | miR-802 | Fresh | 73 | RT-PCR |
| Zhang | 2018 | China | miR-410-3p | Fresh | 82 | RT-PCR |
Associations between miRNAs and the clinicopathological features of prostate cancer
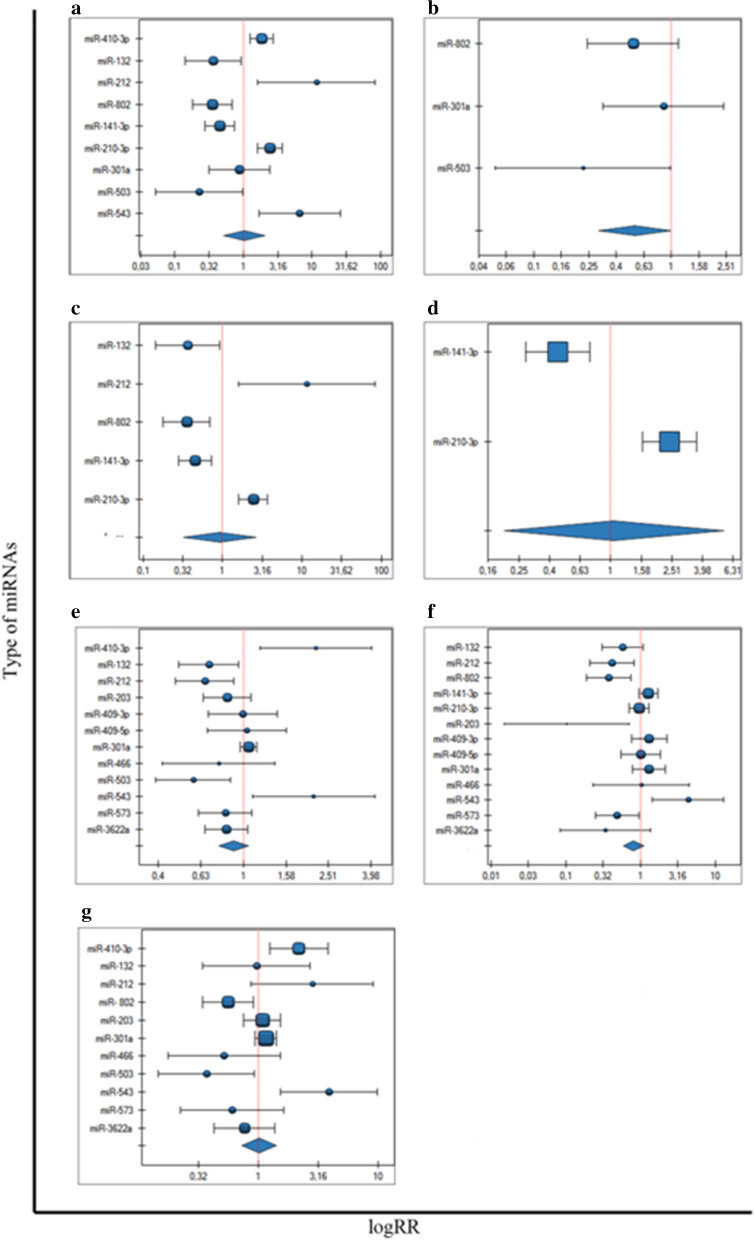
In the current meta-analysis, we checked the association between miRNAs and clinicopathological features of prostate cancer presented in Table 2. For these compilations the Relative Risk (RR) and corresponding 95% Confidence Intervals (95% CI) were calculated.
Table 2.
Associations between miRNAs and clinicopathological features studied in current meta-analysis
| Clinicopathological features | miRNAs | Figures |
|---|---|---|
| Presence of metastasis | ||
| GM | miR-410-3p, miR-132, miR-212, miR-802, miR-141-3p, miR-210-3p, miR-301a, miR-503, miR-543 | Figure 2a |
| LNM | miR-802, miR-301a, miR-503 | Figure 2b |
| DM | miR-132, miR-212, miR-802, miR-141-3p, miR-210-3p | Figure 2c |
| BM | miR-141-3p, miR-210-3p | Figure 2d |
| Total Gleasone scor | ||
| GS > 6 | miR-410-3p, miR-132, miR-212, miR-203, miR-409-3p, miR-409-5p, miR-301a, miR-466, miR-503, miR-543, miR-573, miR-3622a | Figure 2e |
| GS > 7 | miR-132, miR-212, miR-802, miR-141-3p, miR-210-3p, miR-203, miR-409-3p, miR-409-5p, miR-301a, miR-466, miR-503, miR-543, miR-573, miR-3622a | Figure 2f |
| pT stage according to TNM | miR-410-3p, miR-132, miR-212, miR-802, miR-203, miR-301a, miR-466, miR-503, miR-543, miR-573, miR-3622a | Figure 2g |
GM general metastasis, LNM lymph node metastasis, DM distant metastasis, BM bone metastasis, GS Gleason score, pT stage-pathological tumor stage
Association between miRNAs and the metastasis generally
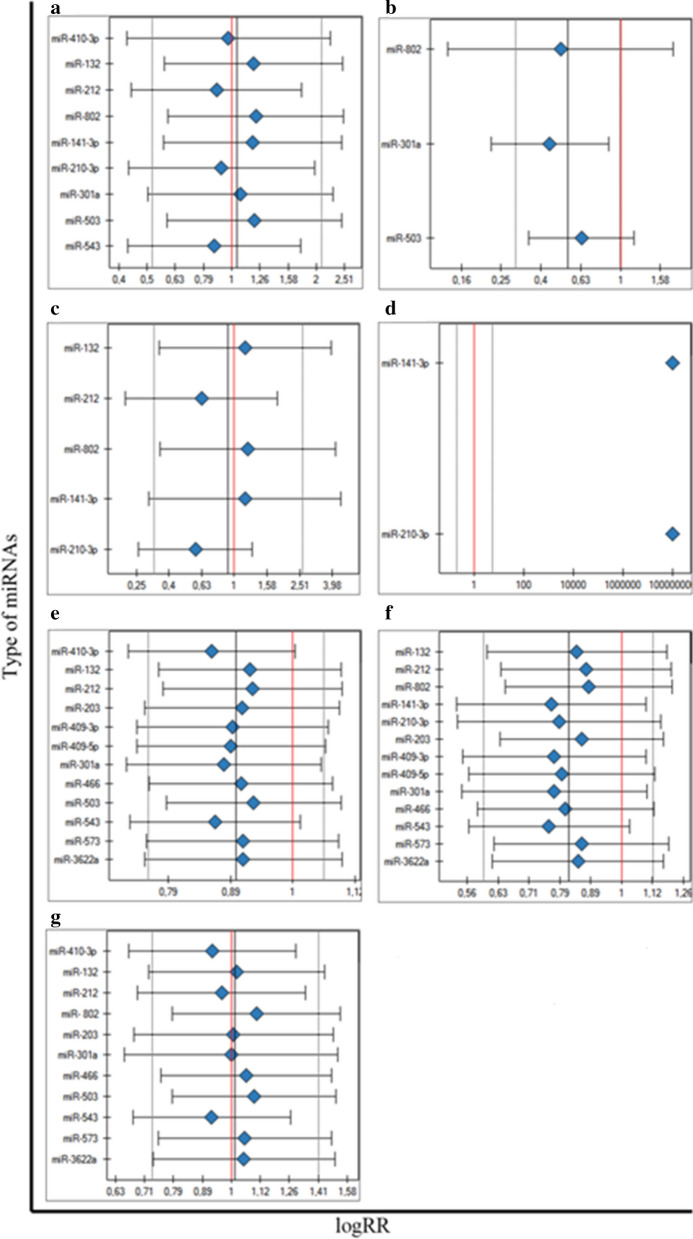
A total of eight articles, involving ten miRNA molecules, were included to analysis of the association between miRNAs expression and the presence of metastasis. The total number of included patients was 881 individuals. The heterogeneity of this analysis was at high level (p < 0.000001, I2 = 87%). Patients with high expression of miR-410-3p, miR-212, miR-210-3p and miR-543 had significantly poorer prognosis compared to their low expression level respectively (RR = 1.85, 95% CI 1.25–2.74, p = 0.002; RR = 11.59, 95% CI 1.61–83.31, p = 0.014; RR = 2.43, 95% CI 1.61–3.67, p = 0.000; RR = 6.60, 95% CI RR = 1.68–25.95, p = 0.005) (Fig. 2a). Furthermore, low expression of other four miRNAs, miR-132, miR-802, miR-141-3p, miR-503 were predicted significantly to tumor progression, compared to high expression of them (RR = 0.36, 95% CI 0.14–0.92, p = 0.033; RR = 0.35, 95% CI 0.18–0.69, p = 0.002; RR = 0.45, 95% CI: 0.28–0.73, p = 0.001; RR = 0.23, 95% CI 0.05–0.99, p = 0.05). In the one analysis with miR-301a no statistically significant association was noticed (Fig. 2a). The clinicopathological details of the group, concerning information about metastases were presented in Table 3.
Fig. 2.
Forest plot analysis
Table 3.
The metastasis presence of included studies
| Author (year) | miRNA | Expression level | Clinicopathological details | |
|---|---|---|---|---|
| Presence of metastasis | p-value | |||
| Du (2017) | miR-543 | Low | 2 | 0.005 |
| High | 12 | |||
| Fu (2016) | miR-132 | Low | 7 | 0.043 |
| High | 6 | |||
| Fu (2016) | miR-212 | Low | 1 | 0.001 |
| High | 12 | |||
| Huang (2017) | miR-141-3p | Low | 36 | < 0.001 |
| High | 16 | |||
| Jiang (2016) | miR-503 | Low | 9 | 0.04 |
| High | 2 | |||
| Nam (2016) | miR-301a | Low | 7 | 0.5260 |
| High | 7 | |||
| Ren (2017) | miR-210-3p | Low | 20 | < 0.001 |
| High | 48 | |||
| Wang (2017) | miR-802 | Low | 15 | 0.002 |
| High | 9 | |||
| Zhang (2018) | miR-410-3p | Low | 17 | 0.001 |
| High | 33 | |||
Sub-analysis 1. Association between miRNAs and the lymph node metastasis
Out of eight published papers with the metastasis patients data the analyses selected three articles, describing miRNAs expression among prostate cancer patients with lymph node metastasis. The total number of engaged patients was 353 individuals, constituting approximately 43% of all PCa patients with lymph node metastasis. The heterogeneity of this sub-analysis was at low level and the heterogeneity was not statistically significant (p = 0.330, I2 = 9.71%). The low expression of miR-503 was associated with the presence of lymph node metastasis (RR = 0.22, 95% CI 0.052–0.989, p = 0.047), compared to high expression of miR-503 (Fig. 2b). The results of others two of three miRNAs were not statistically significant (miR-802 and miR-301a).
Sub-analysis 2. Association between miRNAs and the distant metastasis
From the total eight articles, describing data of metastasis presence, we distinguished four articles, including five miRNAs related to the distant metastasis. This investigated group consisted of 477 individuals, constituting 51% of all patients with metastasis. The heterogeneity analysis for these five miRNAs was at high level (p < 0.000, I2 = 91.59%). The overexpression of miR-212 and miR-210-3p were found to associate with the higher risk of distant metastasis (RR = 11.59, 95% CI 1.61–83.31, p = 0.015; RR = 2.43, 95% CI 1.61–3.67, p = 0.000, respectively), compared to loss of miR-212 and miR-210-3p expression. (Fig. 2c). On the other hand, the downregulation of miR-132, miR-802, miR-141-3p were associated with higher distant metastasis risk (RR = 0.36, 95% CI 0.144–0.924, p = 0.034; RR = 0.35, 95% CI 0.179–0.692, p = 0.002; RR = 0.45, 95% CI 0.277–0.734, p = 0.001) in comparison to the up regulated miR-132, miR-802 and miR-141-3p (Fig. 2c).
Sub-analysis 3. Association between miRNAs and the bone metastasis
Out of eight articles analyzing data of patients with metastases, we distinguished two articles analyzing patients with bone metastases and performed a separate sub-analysis. We decided to include these two articles to sub-meta-analysis apart from the rest types of metastasis. The total number of included patients was 290, which constitutes 35% of all metastatic patients. The heterogeneity test was at high level (p < 0.000, I2 = 96.27%). The results indicated that high miR-210-3p expression level in prostate cancer tissue was significantly associated with the frequent occurrence of bone metastasis (RR = 2.43, 95% CI 1.61–3.67, p = 0.000), compared to low miR-210-3p expression. (Fig. 2d). On the other hand low expression level of miR-141-3p among patients with prostate cancer was associated with the higher risk of bone metastasis occurrence (RR = 0.45, 95% CI 0.27–0.73, p = 0.001) in comparison to the high miR-141-3p expression (Fig. 2d).
Association between miRNAs and the Gleason score
Out of ten articles, we included twelve miRNA molecules to analyze the association between each miRNAs expression level and the total Gleason score > 6. The number of patients was 1448 patients. The heterogeneity test was at middle-high level (p = 0.0001, I2 = 70%). We distinguished five miRNA molecules with RR value statistically significant. We found that elevated expression of miR-410-3p was significantly associated with Gleason score > 6 (RR = 2.19, 95% CI 1.20–4.00, p = 0.01), compared to the loss of miR-410-3p expression level (Fig. 2e). On the other hand, we noticed that decreasing expression of the miR-132, miR-212, mir-503, miR-543 was significantly associated with a total Gleason score > 6 (RR = 0.69, 95% CI 0.49–0.95, p = 0.02; RR = 0.66, 95% CI 0.48–0.90, p = 0.0007; RR = 0.58, 95% CI 0.39–0.81, p = 0.009; RR = 2.14, 95% CI 1.10–4.16, p = 0.02), compared to the higher miR-132, miR-212, miR-503, miR-543 expression level (Fig. 2e).
Among eleven publications, we found 13 miRNA molecules, which were included in the analysis of association with the Gleason score > 7. The number of patients was 1630 individuals. The heterogeneity test was middle-high level (p = 0.00007, I2 = 69.68%). The five of mentioned miRNA molecules were found statistically significant. The analysis showed that low expression level of the miR-212, miR-802, miR-203, miR-543 and miR-573 was significantly associated with the > 7 Gleason score (RR = 0.42, 95% CI 0.21–0.83, p = 0.01; RR = 0.38, 95% CI 0.19–0.75, p = 0.005; RR = 0.1, 95% CI 0.01–071, p = 0.02; RR = 4.33, 95% CI 1.44–13.02, p = 0.009; RR = 0.49, 95% CI 0.25–0.96, p = 0.04) in comparison to the high miR-212, miR-802, miR-203, miR-543 and miR-573 expression (Fig. 2f). The Gleason score details of studied samples were demonstrated in Table 4.
Table 4.
The Gleason score presence of included studies
| Author (year) | miRNA | Expression level | Clinicopathological details | ||
|---|---|---|---|---|---|
| Gleason score | p-value | ||||
| ≤ 7 | > 7 | ||||
| Bucay (2017) | miR-3622a | Low | 89 | 9 | 0.0223 |
| High | 22 | 1 | |||
| Colden (2017) | miR-466 | Low | 57 | 7 | < 0.0001 |
| High | 14 | 2 | |||
| Du (2017) | miR-543 | Low | 18 | 3 | 0.006 |
| High | 8 | 13 | |||
| Fu (2016) | miR-132 | Low | 14 | 19 | 0.028 |
| High | 16 | 8 | |||
| Fu (2016) | miR-212 | Low | 11 | 20 | 0.004 |
| High | 19 | 7 | |||
| Huang (2017) | miR-141-3p | Low | 23 | 48 | < 0.001 |
| High | 54 | 16 | |||
| Jiang (2016) | miR-503 | Low | 11 | 30 | < 0.01 |
| High | 23 | 17 | |||
| Josson (2017) | miR-409-3p | Low | |||
| High | |||||
| Josson (2017) | miR-409-5p | Low | |||
| High | |||||
| Nam (2016) | miR-301a | Low | 268 | 24 | 0.3634 |
| High | 262 | 31 | |||
| Ren (2017) | miR-210-3p | Low | 0 | 22 | < 0.001 |
| High | 0 | 49 | |||
| Saini (2010) | miR-203 | Low | 10 | 1 | |
| High | 11 | 0 | |||
| Wang (2015) | miR-573 | Low | 16 | 18 | 0.044 |
| High | 23 | 8 | |||
| Wang (2017) | miR-802 | Low | 0.004 | ||
| High | |||||
| Zhang (2018) | miR-410-3p | Low | 30 (< 7) | 10 (≥ 7) | 0.006 |
| High | 19 (< 7) | 23 (≥ 7) | |||
Association between miRNAs expression level and pT stage
The eleven miRNA molecules from ten publications have been included to analyze of the association of miRNAs expression and pT stage. The patients' number was 1389 individuals. The heterogeneity test was at middle-high level (p = 0.0003, I2 = 69.53%). The higher risk of the advanced prostate cancer stage was related to the high expression level of miR-410-3p (RR = 2.16, 95% CI 1.23–3.79, p = 0.007) and miR-543 (RR = 3.85, 95% CI 1.52–9.77, p = 0.004), compared to the low miR-210-3p and miR-410-3p expression levels (Fig. 2g). On the contrary, the higher risk of the advanced prostate cancer stage was connected to the miR-802 (RR = 0.55, 95% CI 0.34–0.9, p = 0.02) and miR-503 (RR = 0.37, 95% CI 0.14–0.92, p = 0.03) low expression levels, compared to the high expression level of miR-802 and miR-503 molecules (Fig. 2g). The pathologic tumor stage (pT stage) and details of included samples were shown in Table 5.
Table 5.
The pathological T stage for tumors of included studies
| Author (year) | miRNA | Expression level | Clinicopathological details | ||
|---|---|---|---|---|---|
| Pathological/clinical Stagea | p-value | ||||
| ≤ pT2 | ≥ pT3 | ||||
| Bucay (2017) | miR-3622a | Low | 68 | 22 | 1.000 |
| High | 16 | 1 | |||
| Colden (2017) | miR-466 | Low | 40 | 3 | < 0.0001 |
| High | 13 | 22 | |||
| Du (2017) | miR-543 | Low | 18a | 4a | < 0.001 |
| High | 6a | 14a | |||
| Fu (2016) | miR-132 | Low | 13 | 4 | 1.000 |
| High | 31 | 9 | |||
| Fu (2016) | miR-212 | Low | 23 | 3 | 0.063 |
| High | 21 | 10 | |||
| Jiang (2016) | miR-503 | Low | 27 | 14 | 0.03 |
| High | 35 | 5 | |||
| Nam (2016) | miR-301a | Low | 189 | 103 | 0.2968 |
| High | 175 | 118 | |||
| Saini (2010) | miR-203 | Low | 9 | 2 | |
| High | 10 | 1 | |||
| Wang (2015) | miR-573 | Low | 30a | 11a | 0.003 |
| High | 18a | 23a | |||
| Wang (2017) | miR-802 | Low | 10 | 17 | 0.02 |
| High | 30 | 16 | |||
| Zhang (2018) | miR-410-3p | Low | 29a | 11a | 0.003 |
| High | 17a | 23a | |||
aclinical tumor stage
Regulatory pathways for miRNA-related to EMT program in prostate cancer
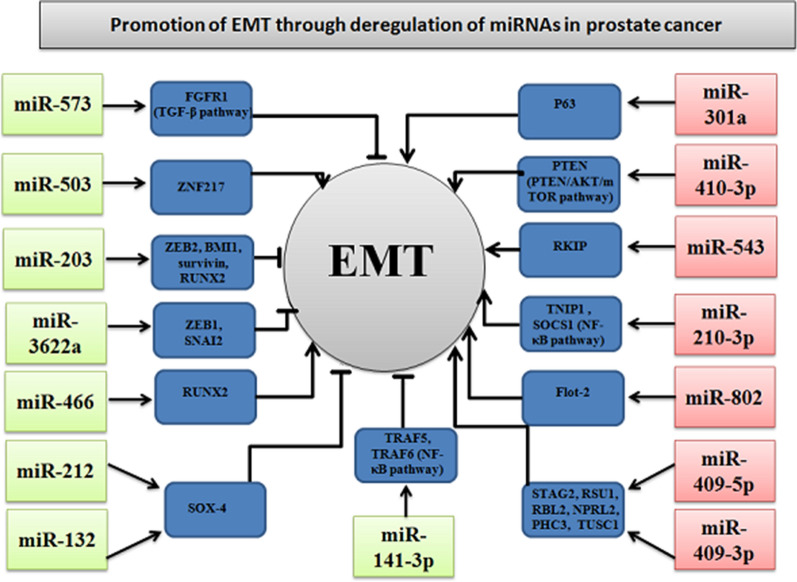
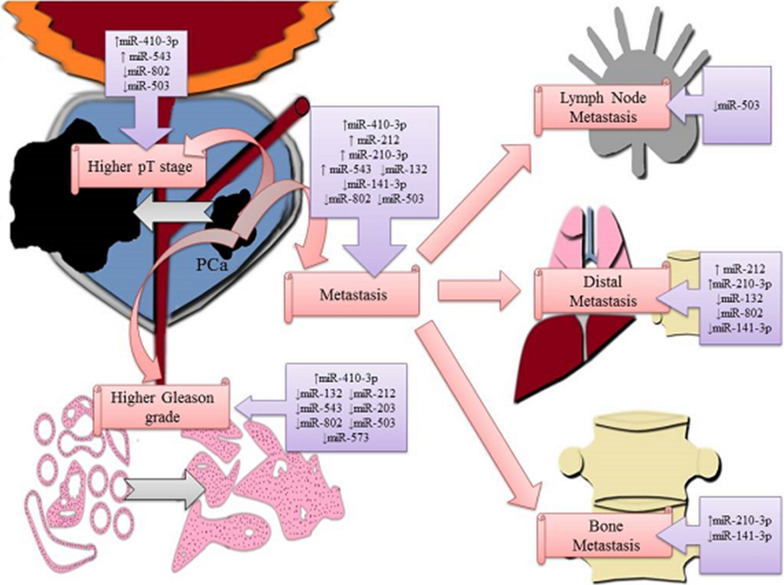
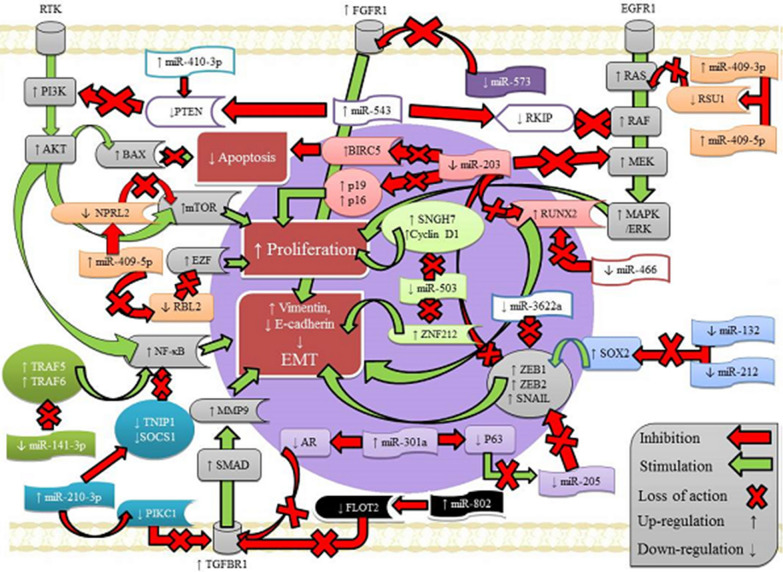
In the current meta-analysis, we obtained the important results for 15 miRNAs, based on primary data from 13 published studies. For 7 miRNAs (miR-410-3p, miR-301a, miR-802, miR-543, miR-409-3p, miR-409-5p, miR-210-3p) of 13 articles (54%), the deregulation was expressed as an overexpression in prostate cancer samples, while for 8 (62%) of 13 published papers the expression was down regulated (miR-573, miR-3622a, miR-466, miR-503, miR-203, miR-141-3p, miR-212, miR-132). The direct targets of specific miRNAs and their expression alterations in a prostate cancer were presented in the Fig. 3. The associations between specific deregulation of miRNAs expression and following parameters: clinicopathological features, including general, lymph node, distant or bone metastasis, the both Gleason scores > 6 and > 7 as well as pT stage were demonstrated in Table 6. For better understanding aforementioned dependencies, the expression signatures for individual clinicopathological features were demonstrated in Fig. 4. The various mRNA targets of dysregulated miRNAs and their signal transduction pathways in prostate tumor cells were demonstrated in Fig. 5. The presentation of key regulatory molecular mechanisms of analyzed miRNAs involved in progression to metastatic disease supports the understanding of the underlying miRNA functions in prostate cancer cells.
Fig. 3.
Targets of specific miRNAs-related to EMT program in prostate cancer
Table 6.
Association between miRNAs expression and clinicopathological features
| GM | LNM | DM | BM | GS > 6 | GS > 7 | pT stage | |
|---|---|---|---|---|---|---|---|
| miR-132 | ↓ | ↓ | ↓ | ||||
| miR-141p | ↓ | ↓ | ↓ | ||||
| miR-503 | ↓ | ↓ | ↓ | ↓ | |||
| miR-212 | ↓ | ↓ | |||||
| miR-573 | ↓ | ||||||
| miR-210-3p | ↑ | ↑ | ↑ | ||||
| miR-410-3p | ↑ | ↑ | ↑ | ||||
| miR-543 | ↑ | ↑ | |||||
| miR-203 | ↓ |
GM general metastasis, LNM lymph node metastasis, BM bone metastasis, GS > 6 Gleason score > 6, GS > 7 Gleason score > 7, pT stage pT stage according to the TNM system, ↑ overexpressed miRNA, ↓ downregulated miRNA
Fig. 4.
MicroRNA expression signatures related to tumor stage, Gleason grade and progression to metastatic prostate cancer
Fig. 5.
Signaling pathways scheme depicting prostate cancer progression-related targets of dysregulated miRNAs in tumor cells
Many previous studies have indicated that miRNAs play a crucial role in various biological processes, encompassing development, proliferation, differentiation, cell fate determination, apoptosis, angiogenesis, adhesion, metastasis, signal transduction, organ development, hematopoietic lineage differentiation, host-viral interaction and tumor genesis [34–40]. The known biological functions of analyzed miRNAs in the cancerous tissues have been presented in Table 7.
Table 7.
Biological functions of discussed miRs in cancer
| Name of microRNA | Genomic locations | Biological functions in cancer | Ref. |
|---|---|---|---|
| miR-3622a | 8p21.1 | miR-3622 regulates cancer cells proliferation, progression, apoptosis, G0/G1 cell cycle arrest, migration and invasion abilities of cancer, EMT, metastasis | [23, 41–44] |
| miR-466 | 3p23 | miR-466 induces proliferation, migration, invasion, angiogenesis cell cycle arrest, apoptosis in cancer cells, EMT and metastasis | [25, 45, 46] |
| miR-543 | 14q32.31 | mir-543 mediates cell proliferation and tumor growth, apoptosis, the cell cycle, migration, invasion, EMT and metastasis | [30, 47–53] |
| miR-132 | 17p13.3 | miR-132 regulates proliferation of cancer cells, colony formation, tumor growth, migration and invasion of cancer cells, the G1/S phase transition of the cell cycle, EMT in cancer cells, apoptosis in cancer cells | [54–58] |
| miR-212 | 17p13.3 | miR-212 mediates the proliferation and invasion of cancer cells, EMT and metastasis | [59–62] |
| miR-141-3p | 12p13.31 |
miR-141 regulates proliferation, tumor growth, migration, EMT, chemosensitivity, apoptosis, angiogenesis, oxidative stress response, cellular motility and control “stemness” miR-141–3p regulates proliferation, tumor growth, migration, and invasion of cancer cells, cancer cell progression, EMT and metastasis |
[33, 63–70] |
| miR-503 | Xq26.3 | miR-503 regulates cell proliferation, tumor angiogenesis, cell growth and EMT in cancer cells | [71–77] |
| miR-409-3p | 14q32.31 | miR-409-3p mediates tumorigenesis, tumor growth, invasion, EMT and metastasis of tumor cells | [32, 78–81] |
| miR-409-5p | 14q32.31 | miR-409-5p mediates tumorigenesis, EMT and bone metastasis | [32, 81] |
| miR-301a | 17q22 | miRNA-301a regulates tumor cell invasion, migration, cell proliferation, apoptosis and enhancing chemosensitivity, EMT and metastasis | [82–84] |
| miR-210-3p | 11p15.5 | miR-210-3p regulates cell survival, stem cell differentiation, angiogenesis, DNA damage repair, mitochondrial metabolism, immune response, EMT and metastasis | [28, 63, 85–89] |
| miR-203 | 14q32.33 | miR-203 regulates tumor proliferation, invasion, migration, apoptosis, EMT and metastasis | [90–93] |
| miR-573 | 4p15.2 | miR-573 mediates cell proliferation, tumor growth, apoptosis, angiogenesis, EMT and invasion of cancer cells | [94–96] |
| miR-802 | 21q22.12 | miR-802 modulates proliferation, tumor growth, apoptosis, migration, invasion, EMT and metastasis of cancer cells | [29, 97–100] |
| miR-410-3p | 14q32.31 | miR-410-3p mediates cell proliferation, invasion, migration, EMT, apoptosis, chemosensitivity and autophagy | [21, 101–103] |
Publication bias
The high heterogeneity level of data of assembled miRNAs could be related to the variables selected in each study. The most frequently heterogeneity sources of variance in meta-analyses are different research methodologies, various patient races, various research samples and methods of sample collection or storage, insufficient data, the small study groups, different analysis periods (follow-up time), various indicators and methods of estimating results. In order to evaluate disturbing factors we performed sensitivity analysis.
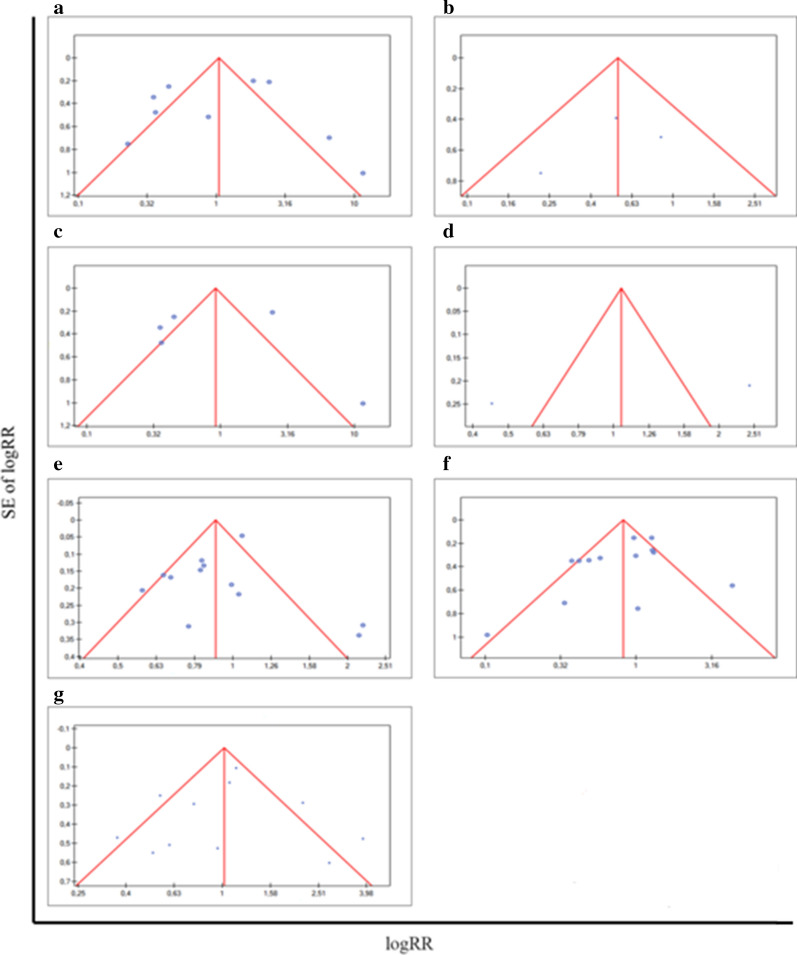
We performed Begg’s funnel plot and Egger’s test to judge potential publication bias of completed analysis. Significant heterogeneities (p < 0.05, I2 > 50%) were found in a miRNAs and metastasis analysis. The heterogeneity test performed for the analysis of the association of miRNAs and the general metastasis indicated high level (I2 = 87.99%) and was statistically significant (p < 0.00002). I2 > 75% indicates a high heterogeneity of the study, which could be disturbed by different types of metastasis. The main results for pooled RRs for meta-analysis were recapitulated in Table 8. It was right to consider a more homogenous group to meta-analysis in the way of excluding miRNAs, which lead to perturb a heterogeneity of study. In order to reduce the risk of heterogeneity and better presenting of the results, we decided to repeat the analysis formed subgroups based on metastasis type: lymph node metastasis, distant metastasis and bone metastasis. The associations of miRNAs and both presence of distant metastasis and bone metastasis revealed the high level of heterogeneity test and the results was statistically significant (I2 = 91.59%, p < 0.000001; I2 = 96.27%, p < 0.000001, respectively). However, the association between miRNAs expression and lymph node metastasis noticed a low level of heterogeneity test at I2 = 9.71% and was statistically insignificant. Others compilations indicated a moderate heterogeneity level: I2 = 70.09%, p = 0.000125 for GS.6; I2 = 69.68%, p = 0.000085 for GS > 7; I2 = 69.53%, p = 0.000293 for pT stage. The Egger’s test, counting the asymmetry of conducted analysis, showed statistically non-significant results for all compilations. The funnel plots presenting publication bias were shown in Fig. 6.
Table 8.
Main results of pooled RRs in the meta-analysis
| Comparisions (miRNA groups) | Heterogeneity test | Summary RR (95% CI) | Asymmetrical distribution | Studies | |||
|---|---|---|---|---|---|---|---|
| Q | p-value | I2 (%) | b Egger coefficient | p-value | |||
| General metastasis | 66.60 | < 0.000001 | 87.99 | 1.05 (0.52, 2.09) | − 0.86 | ns (0.70) | 9 |
| Distant metastasis | 47.58 | < 0.000001 | 91.59 | 0.92 (0.32, 2.61) | − 0.82 | ns (0.86) | 5 |
| Lymph node metastasis | 2.21 | ns (0.33) | 9.71 | 0.54 (0.30, 1.00) | − 1.80 | ns (0.65) | 3 |
| Bone metastasis | 26.83 | < 0.000001 | 96.27 | 1.05 (0.84, 5.49) | NA | NA | 2 |
| Gleason score > 6 | 36.78 | 0.000125 | 70.09 | 0.90 (0.77, 1.06) | − 0.71 | ns (0.46) | 12 |
| Gleason score > 7 | 39.58 | 0.000085 | 69.68 | 0.82 (0.60, 1.13) | − 1.55 | ns (0.15) | 13 |
| pT stage | 32.81 | 0.000293 | 69.53 | 1.02 (0.73, 1.41) | − 0.31 | ns (0.77) | 11 |
Fig. 6.
Funnel plots for publication bias
Sensitivity analysis
We performed sensitivity analysis to establish the impact of single miRNA studies on the stability of achieved results through removing individual miRNA study, in a sequential manner. For the analysis of association between miRNAs expression level and the general metastasis, we noticed that three studies with miR-212, miR-210-3p and miR-543 had the largest effect on the results, but each p-value was statistically non-significant. The rest of the studies showed the RR value close to one or greater than one and constituted stable and robust results. For the comprehensive analysis of individual miRNAs expression and specific metastasis types, the sensitivity analysis demonstrated disorder results for miR-301a, concerning the lymph node metastasis, with the statistically significant p-value (p = 0.018181) as well as miR-212 and miR-210-3p, concerning the distant metastasis, but the p-value was not statistically significant. The RR values of the rest engaged studies of mentioned two analyses were close to each other. For GS > 6, the sensitivity analysis showed two studies with miR-410-3p and miR-543, disturbing the association analysis, but the p-value indicated statistically non-significant results. The rest of RR value of miRNA studies was close to one and stable. Similarly, for pT stage, the sensitivity analysis showed three studies (with miR-410-3p, miR-212, miR-543) with RR value, deviating from the rest studies, but the p-value revealed statistically non-significant results. The diagrams illustrating sensitivity analysis were included in Fig. 7.
Fig. 7.
Sensitivity analysis for meta-analysis groups and subgroups
Discussion
MicroRNAs are molecules which show promise as potential markers of poor prognosis among patients with malignancies. The latest advances in prostate cancer research showed diagnostic, prognostic and treatment utility of variety miRNA molecules measured in serum, plasma, ejaculate, tissue or urine [104–108]. Nonetheless, our study describes the significance of miRNAs involved in the EMT program for the first time, incorporating 1608 prostate cancer patients.
Cancer metastases are a leading cause of cancer mortality and accounts for approximately 90% of deaths due to cancer [109]. However, appropriate diagnostic techniques for predicting progression potential have still not been discovered. Our results provided by 13 tissue-based studies for 7 overexpressed and 8 down regulated miRNAs revealed 9 miRNAs with the highest association with metastatic stage of PCa. The current meta-analysis suggested that miR-210-3p and miR-141-3p had particularly notable significance in a formation of metastasis. Overexpressed miR-210-3p and down regulated miR-141-3p have been found to be associated with the presence of general metastasis, distant metastasis as well as bone metastasis.
MiR-210-3p has been widely recognized as an oncogene, engaged in tumor progression process. MiR-210-3p was documented as up regulated in many cancers: breast cancer [110], lung cancer [111], pancreatic cancer [112], head and neck cancer [113]. In breast cancer, miR-210-3p was recognized in rapid formation of distant metastases [114–116]. Similarly, miR-210-3p expression was found to be elevated in bone metastatic PCa patients compared to non-metastatic PCa patients. Additionally, it was found that miR-210-3p silencing has significantly attenuated bone metastasis formation in PC-3 cells in vivo [28]. As a consequence, it is likely that through the assessment of miRNA levels, metastasis types could be differentiated. Our meta-analysis revealed characteristic miRNAs signatures for each metastasis type. Based on presented results a downregulation of miR-503 seems to be specific for lymph node metastasis while a upregulation of miR-210-3p as well as a downregulation of miR-132 and miR-141-3p are associated to presence of distant metastasis. The signature of aforementioned miRNAs could be potentially used to estimate a risk of metastasis development at an early stage of PCa followed by a more radical treatment procedure in selected cases. On the other hand, the miR-141-3p has been found acting as a tumor-suppressing miRNA, inhibiting the cancer cells progression via directly targeting of tumor necrosis factor receptor-associated factor 5 (TRAF5) in colorectal cancer tissues and cell lines [117]. Similarly, at PCa study, miR-141-3p has been found down regulated, acting as a tumor suppressor that targets TRAF5 and TRAF6 [33].
In the current meta-analysis it was shown that the formation of metastasis overall has been significantly associated with increased expression of miR-410-3p and miR-543. The recent study of Zhang et al. has shown that the elevated expression of miR-410-3p affects a downregulation of phosphatase and tensin homolog deleted on chromosome ten (PTEN) and results in an activation of AKT/mTOR signaling pathway. They have also shown that miR-410-3p plays a crucial role as an oncogene in PTEN/AKT/mTOR pathway [21]. It is possible that its upregulation could promote a disease progression and a metastatic ability of prostate cancer due to deactivation of PTEN protein, a natural inhibitor of PI3K/AKT/mTOR pathway. Similar mechanism may be associated with increased miR-543 level. In colorectal cancer (CRC), miR-543 has been found acting as an oncogene and promotes tumor progression [47]. Moreover, MiR-543 has been significantly up regulated in metastatic prostate cancer tissues [30]. Sun et al. have revealed that PTEN is a direct target of miR-543 [47]. In turn, PTEN has been considered as a tumor suppressing gene, which is strongly involved in the apoptosis process [118]. In the another study, Liu and co-workers have noticed that rising expression of miR-543 may lead to a downregulation of PTEN and inversely, the inhibition of miR-543 could activate PTEN in CRC cells [51]. In many previous studies have been shown that the loss of PTEN gene can increase the PI3K/AKT signal transduction pathway, leading to the tumor progression and formation of metastases [119, 120].
Furthermore, MiR-543 has been also described as a pivotal modulating factor of tumor growth and metastasis through binding to Raf kinase inhibitor protein (RKIP) in PCa. Previous studies have suggested that RKIP is responsible for inhibition of PCa progression by blocking MAP kinase (MAPK) signaling pathway and activation of NF-ƙB factor through G-protein regulating [121, 122]. Li and colleagues in their study at gastric cancer (GC) have demonstrated that miR-543 enhances ability to cell proliferation and formation of colony, what in consequence lead to a progression to the S phase, by directly targeting histone deacetylase sirtuin 1 (SIRT1) [49]. In the same study, the authors have shown that miR-543 molecule negatively modulates the SIRT1 expression in GC cells. Moreover, themiR-543 was positively associated with such clinicopathological parameters as size of the tumor, clinical grade, TNM stage and metastasis to the lymph nodes in gastric cancer patients [49]. Additionally, Haga and co-workers have presented seven miRNAs: miR-300, miR-382, miR-494, miR-495, miR-539, miR-544 and miR-543 in the imprinted DLK1-DIO3 region, which cooperatively regulates the EMT process, by repressing signal transduction network, including TWIST1, BMI1, ZEB1/2, and miR-200 family miRNAs [123]. The epigenetic silencing of miRNAs cluster that appears through hypermethylation of upstream CpG islands in human ductal carcinomas lead to initiate the EMT process [123].
On the other hand, we observed also in current meta-analysis that microRNA molecules might be closely related to a specific type of metastases. The decreased miR-503 expression could correspond with a formation of metastasis generally and lymph node metastasis specifically, while the decreased expression of the miR-132 seems to be associated with the development of metastasis overall as well as with the presence of distal metastasis. These observations may suggest a possibility of determining a risk of lymph node metastasis or distant metastasis development via changes in miR-503 and miR-132 level. MiR-503 has been found to be involved in formation of a malignant phenotype in a wide spectrum of cancers. Using the RT-qPCR, it has been demonstrated the elevated expression level of miR-503 in esophageal squamous cell carcinoma (ESCC). Also, the overexpression of miR-503 was found in correlation with following clinicopathological features: TNM stage, tumor differentiation and lymph node metastasis. Summarizing this study, the authors revealed that miR-503 represses ESCC progression by induction of autophagy, regulated in the PKA/mTOR signaling pathway [124]. Therefore, a decrease in miR-503 level leads to tumor proliferation and metastasize. In human non‐small cell lung carcinoma (NSCLC), miR-503 was observed as a tumor-suppressive miRNA, which was inversely associated with patient overall survival. In turn, upregulation of either PI3K p85 or IKK-β, direct targets of miR-503, guaranteed fractionally restoring of malignant phenotype of cancer cells [125]. Interestingly, in A549/CDDP non-small cell lung cancer cells in comparison to the parental A549 cells, deregulation of miR-503 expression was found to decrease the resistance on cisplatin of NSCLC cells [126]. These findings may have important significance in case of chemotherapy failure due to cancer drug resistance. However, in current work we point out a possibility of miR-503 implementation to predict progression to the lymph node metastasis stage, specifying their potential between miR-503, miR-802 and 301a.
The presence of lymphovascular metastasis constitutes a formidable foundation for the possibility of further extending metastases and forming the tumor cells foci, located at distant body parts. Hence, in the future perspective, a prediction of metastasis development and a clinical classification of metastasis types via fluctuations in microRNA molecules expression levels is justified. Predicting lymph node metastasis by evaluation of miR-503 expression possess a valuable potential in particular for the possibility to inhibit spreading of tumor cells to distant body parts.
Contrarily, miR-132 can play a dualistic role in cancers [127]. In this meta-analysis, miR-132 was found to be associated with both general metastasis and distant metastasis The results of latest studies may confirm an oncogenic role of miR-132 in cancer. Wong and co-workers have found 24-upregulated miRNAs, including miR-132, in tongue squamous cell carcinoma [128]. Liu et al. have revealed that miR-132 was down-regulated in both hepatocellular cancer (HCC) tissues and HCC cell lines and the same miR-132 was responsible for a suppression of tumor development and blocking of invasiveness. Additionally, miR-132 can directly target PIK3R3 and controls the AKT/mTOR signal transduction pathway in HCC [58]. MiR-132 may act as a tumor suppressing miRNA in non-small lung cancer [56] and ovarian cancer [129]. In PCa the loss of miR-132 was found to be in correlation with poor clinical outcome. MiR-132 deregulation leads to decreased a cell-to-cell adhesion and to cell death inhibition, which is necessary for circulating tumor cells to adhere to foreign organ areas. Formosa et al. have observed that the CpC island hypermethylation seems to be responsible for silencing of miR-132 [130].
Such highly heterogeneous tumor as PCa creates significant diagnostic challenges and hinders the ability to predict disease progression. Exploiting the expression alterations of miRNAs specific for kind of metastasis may provide opportunity to early prediction of prostate cancer progression. In this meta-analysis, we want to point out the potential of panel with 6 miRNAs for monitoring disease progression, directed to metastasis occurrence. Our findings revealed that the increased expression of miR-410-3p, miR-210-3p, miR-543 and the decreased expression of miR-132, miR-141-3p and miR-503 have been significantly associated with the appearance of metastases in progress of PCa.
The second aim of our study was to analyze a relationship between miRNAs in PCa and a Gleason score. The Gleason scoring system constitutes an especially important prognostic factor associated with the growing aggressiveness of neoplastic changes in the prostate. The increased Gleason grade has been connected inter alia with pathologic stage, tumor size, margin status, progression to metastatic form and patient survival [131]. Total Gleason score is one of the main factors involved in making treatment decisions, together with pathological stage and pre-PSA level [132].
In our meta-analysis we decided to compare the miRNAs importance in the set of “Gleason score above 6” and in the set of “Gleason score above 7”. Aberrant miR-410-3p, miR-132, miR-212, miR-503 are separately the most associated with total Gleason score > 6 and abnormal expression of miR-212, miR-203 and miR-573 are individually the most associated with Gleason score > 7 in PCa tissue. MiRNAs molecules most integrated with both Gleason scores above 6 and 7 is miR-212. The future implementation of diagnostic tests to capture the variations in miRNAs expression among prostate cancer patients limiting to Gleason score, suggests the opportunity of monitoring the patient’s condition and disease progression at early cancer stage, excluding need to resection of prostate gland or radiotherapy.
The miR-212 was found to be upregulated in pancreatic ductal adenocarcinoma in comparison to the health tissue. In the current study, it has been shown the positive correlation between deregulation of miR-212 and TNM stage, lymph node metastasis, vessel invasion, size of the tumor and the overall survival time. Latest, it has been found that in hypoxic conditions miR-212 aberration was correlated with the hypoxia-inducible factor (HIF-1α) in vivo and in vitro [133]. In turn, in cervical cancer tissues and cell lines, miR-212 was found outstandingly down regulated and promoted proliferation and invasion of cancer cells. Using the Western blot tool the authors have indicated that miR-212 upregulation notably blocked the transcription factor 7-like 2 (TCF7L2) protein expression level [134].
Furthermore, miR-212 contributes in the miR-212/132 cluster in human lung cancer [135]. Interestingly, the current study also showed an association between miR-132 level and Gleason score > 6. Jiang et al. have suggested that overexpression of miR-212/132 cluster remarkably blocks the growth and invasiveness of tumor cells in A549 and H1299 lung cancer cell lines [135]. Additionally, miR-212/132 upregulation leads to activation of cell cycle arrest at the G1/S phase transition via regulation of p21 and cyclin D1 expression [135]. Upregulation of miR-132/212 has been found in drug resistant breast tumors and cell lines. Moreover, the miR-132/212 has been shown responsible for regulation of drug accumulation through blocking the PTEN expression levels in vitro. Also, downregulation of PTEN has been found inversely associated with miR-132/212 expression in drug resistance breast cancers. Upregulation of miR-132/212 cluster was partially connected with transactivation via the NF-ƙB transcription factor and this mechanism may have importance in acquisition of drug resistance [136]. Additionally, current statistical analysis showed that another aforementioned miRNAs, miR-410-3p and miR-503 was found to be in association with Gleason score value > 6, which demonstrates their potential prognostic role at an early stage of prostate cancer development.
MiR-573 and miR-203 was found to be associated only with Gleason score > 7, therefore we consider that these molecules exhibit a lower prognostic value for predicting EMT. However, miR-573 and miR-203 can be used to predict Gleason score ranges from 7 to above. Wang et al. have investigated that upregulation of miR-573 may be stabilized by miR-573 mimic, leading to inhibition of proliferation and progression in melanoma cells [137]. On the other hand, Wang and other authors have revealed that downregulation of miR-573 in PCa is precisely associated with the Gleason score and cancer-related mortality (p = 0.41). The latest study showed that miR-573 could regulate the activity and function of Fibroblast growth factor receptor 1 (FGFR1) in response to fibroblast growth factor 2 (FGF2). Additionally, in the same study, transcription factor GATA3 was indicated as an enhancer of miR-573 expression, causing downregulation of FGFR1, EMT and invasion of PCa cells [22].
MiR-203 has been found to be a tumor suppressing miRNA in some cancers: hepatocellular cancer [138], breast cancer [139, 140], esophageal cancer [141], leukemia [142], glioma [143], bladder cancer [144], leading to inhibition EMT by targeting various genes and downregulation of signal transduction pathways, thus is involved in cancer progression (tumor proliferation and invasion). However, its role in some cancers has been found different. MiR-203 can function as an oncogene in ovarian cancer [145]. In SKOV3 and OVAR3 ovarian cancer cell lines, high expression of miR-203 leads to restrain of EMT process by linking to BIRC5 and causes its downregulation and attenuates the TGF-β signal transduction pathway [92, 146]. Chen and Ding et al. have investigated the effect of elevated miRNA expression on PC-3M prostate cancer tumor cells behavior and they found that downregulation of miR-203 was connected with adriamycin (ADM) resistance, thus elevation of miR-203 repress tumor cells proliferation, enhance apoptosis and reduce ADM resistance through influencing on MAPK kinase 1 (MEK1) expression [147]. MEK1 is responsible for phosphorylation of the Tyr/Thr residue on ERK protein and leads to induction of ERK/MAPK transduction pathway. In turn, excessive stimulation of ERK/MAPK pathway intensifies progression of tumor cells [147].
Finally, we analyzed the association between miRNAs and the pT stage of PCa. The process of determining tumor stages among prostate cancer patients relies on evaluation of probability of tumor spread before including the individual treatment [148]. The increased expression of both miR-410-3p and miR-543 as the same as decreased expression of miR-503 have a significant relationship with rising probability of tumor extending and higher T stage at the time of diagnosis. Our results may be useful for prediction of prostate cancer progression at an early stage. Forasmuch as further research is needed in the prostate cancer progression area, it is important to be prudent in devising far-reaching conclusions. However, it was noticed that progression to a higher pT stage is associated with the aberration of 3 miRNA levels: miR-410-3p, miR-543 and miR-503 involved in invading surrounding tissues and metastasizing. Du et al. at their study on CRC, using gene chip technology grouped inter alia by stage, have proven that microRNA expression profiles were significantly different at various CRC stages, thereby confirming that miRNAs are engaged in a cancer development and in particularly are significant at early diagnosis or cancer prognosis [149]. Concluding, the probability of cancer progression to its metastatic stage can be assessed at an early stage of tumor development. MiR-410-3p play a different role in various cancers. Zhang et al. have noticed that increased expression of miR-410-3p repress proliferation and invasion process in the breast cancer cells [150]. These authors also displayed that transcription factor Snail is a direct target of miR-410-3p and in this way miR-410-3-p inhibits an EMT process. MiR-410-3p has been demonstrated to inhibit the invasion and migration of rhabdomyosarcoma cells through inhibiting EMT [151]. Wang and other authors suggested that miR-410 function in CRC may be connected to negative regulation of DKK-1 through Wnt/β-catenin pathway [152]. MiR-410-3p plays a significant role as an oncogene in PCa via PTEN/PI3K/AKT pathway. Downregulation of miR-410-3p leads to an activation of PTEN, which is a crucial modulator of the PI3K/AKT signaling pathway [21]. Similarly, the role of miR-543 in tumor progression and metastasis were confirmed in many earlier publications [52, 153, 154]. It has been also reported that miR-543 shows outstanding overexpression in the metastatic PCa cell lines compared to the normal cells. Additionally, Raf kinase inhibitor protein (RKIP) was found to be a direct target of miR-543. Upregulation of miR-543 leads to decrease in expression level of RKIP and promotes proliferation and metastasis in PCa cells. [30]. Previous studies have revealed that described molecules are undeniably involved in initiation EMT transition, therefore they are ideal candidates for metastasis risk prediction.
Diagnostic, prognostic and therapeutic aspect of analyzed miRNAs
The revolution in the diagnosis of prostate diseases was measuring PSA level in blood samples, however it is questionable due to the lack of association between PSA level and prostate cancer stage, which may result in overdiagnosis and overtreatment. Currently, many scholars put immense effort into identifying new biomarkers to improve the diagnosis and prognosis accuracy of PCa entities. Previous studies have indicated that specific miRNA profiles were able to distinguish cancerous and non-cancerous tissues [155], incoming the potentiality of miRNAs in early cancer detection, prognosis and monitoring of treatment response.
Tissue biopsies represent a gold standard in prostate cancer patients’ diagnosis. Nevertheless, tissue sampling technique is susceptible to misdiagnosis, whereas a negative outcome cannot completely exclude presence of cancer foci. Hence, the researchers are looking for a better diagnostic tool. miRNA molecules have multiple advantages in diagnostic or treatment approach, for example, RNA has a higher specificity and its expression alterations are characteristic for individual disease. The expression of miRNA in cancer cells is a process during which can be observed changes that appear over disease development. Additionally, in comparison of tissue-based miRNAs with detection of miRs in bodily fluids, the second one mentioned can be associated with many factors, such as age, diet or circadian rhythms [156]. miRs are less subjected to degradation due to their small size. Additionally, miRs possess great stability during FFPE tissue processing since they are encapsulated in exosomes [156].
Current research state constitutes that non-coding RNA, such as miRNAs, has great potential for PCa diagnosis. An individual gene can be targeted by various miRNAs, which accelerate atypical cell growth and provoke cell death [157]. Furthermore, miRNAs can manage cancer-related processes, such as EMT or metastasis. MiRNAs can be measured both by tissue-based biopsy or in various bodily fluids [158]. Aghdam and colleagues indicated that tissue miRNAs are appropriate for diagnosis and staging of malignancies as well as minimize the necessity for numerous biopsies to determine the appropriate diagnosis [158]. Even if the miRNA detection on tissue-based biopsies shows multiple benefits for prostate cancer patients, it is not yet applied in present clinical practice.
Certain miRNA molecules described in current meta-analysis have been studied previously for diagnostic and prognostic potential in prostate cancer patients. MiR-141 in association with miR-151-3p and miR-16 was found to have a sensitivity of 84% and a specificity of 96% regarding mCRPCa detection from liquid samples [159]. Agaoglu and colleagues presented that circulating miR-141 in compilation with miR-21 and miR-221 have potential to differentiate PCa patients in metastatic stage from those with local advance stage [160]. Other authors have shown that the combination of miR-141-3p with miR-21 and miR-375 presented a sensitivity of 93% and a specificity of 63% during prediction of PCa in serum samples [161]. MiR-141 is a widely researched molecule in various cancers, useful at each stage of diagnosis and prognosis for prostate cancer patients.
Also miR-301a was found to be a valuable diagnostic and prognostic biomarker for prostate cancer disease. The study performed on 28 prostate cancer patients and 13 controls as well as 40 radical prostatectomy cases indicate the possible correlation between miR-301a detected in serum and Gleason score [162].
MicroRNAs are validated as necessary tools in designing biomarker panels for identification prostate malignancies primarily in early but also advanced stages. Regulation of aberrant miRNA expression will repress the targets responsible for progression to higher prostate stage. Presently, TNM staging along with Gleason score and pretreatment PSA levels are utilized to predict the outcome for prostate cancer patients. The microRNA analytes in current meta-analysis have potential for higher accurate prostate cancer staging and developing therapy schemes. Additional advantage for miRNA diagnostic value in prostate cancer is the fact that miRs are situated at fragile chromosomal sites corresponding with tumor hotspots [12].
The role of miRNAs in prostate malignancies is not only limited to diagnosis or prediction and prognosis. MiRNA molecules can also function as targets or therapeutic agents. MiRNA molecules can regulate multiple target genes, therefore alterated expression constitutes a potential therapeutic value for the balance of gene expression [163]. Aside from direct application of miRNA as a valuable therapeutic option, miRNAs can facilitate selection of the best individual treatment scheme and predict response to a personal therapy [158]. The individual patients' response to a treatment application, such as radical prostatectomy, radiotherapy, testosterone suppression and hormone therapy, chemotherapy and immunotherapy is different [158, 164]. Intriguingly, miRNA can be applied as adjuvants acting as an enhancer of tumor sensitivity to therapy [158].
Zedan and other authors have investigated the relationship between circulating miR-141 and miR-375 and treatment outcome in metastatic castration resistant prostate cancer (mCRCP), where 40 patients were treated with docetaxel and 44 with abiraterone [165]. The results of the mentioned study showed prognostic importance of miR-141 and miR-375 in treatment response monitoring among mCRPC patients. Intriguingly, Gonzales and colleagues noticed a strong correlation between miR-141 levels and chemotherapy response in patients with confirmed metastatic stage of prostate cancer. Furthermore, they proved that multivariate panel of directional alterations in PSA, circulating tumor cells (CTCs) and miR-141 had higher sensitivity of 78.9% in clinical outcome prediction [166]. Saini and other authors have studied miR-203 in advanced metastatic prostate cancer and revealed that miR-203 may act as an “antimetastatic” miRNA at numerous steps in the metastatic process by repression of prometastatic targets. Hence, miR-203 has been recognized as an attractive target for patient therapy in advanced prostate disease [27]. The prognostic value of miR-141 and miR-409-3p was demonstrated by Nguyen and other authors, investigating the differential expression of 669 miRNA prostate cancer serum. They confirmed that miR-141, miR-375 and miR-378 increase-depend manner with progression to higher prostate cancer stage, while miR-409-3p was marked to be overexpressed in high-risk group compared to low-risk group, but significantly reduced in the metastatic CRPC, which probably was associated with the androgen deprivation therapy [167]. Watahiki and colleagues proposed miRNA panel, including miR-141, miR-152 and miR-423-3p significantly deregulated in metastatic CRPC compared to localized PCa emerged from 742 miRNAs studied in plasma samples, demonstrating AUC of 0.944. Additionally, the researchers confirmed that miR-141 together with miR-151-3p, miR-152 and miR-423-3p was linked with poor outcome and higher Gleason score. Furthermore, miR-141 and miR-152 was able to detect a high probability of recurrence among patients after radical prostatectomy [159]. In contrast to serum-based miRNA studies, Porkka et al. have profiled 319 miRNA molecules in nine PCa and compared to them four BPH tissues, shown inter alia miR-210 together with other 7 miRNAs up regulated and 22 miRNAs down regulated [168]. Interestingly, Srivastava et al. have shown 30 miRNAs, including miR-210 and miR-212 down regulated and three up regulated in 40 FFPE tissue specimen blocks, comparing PCa tissues to adjacent normal tissue [169].
Clinical relevance of miRNA molecules constitutes the basis to widespread research in the therapeutic context. Therefore, investigators are underway to explore systems balancing miRNA expression levels, onco-miRNA antagonizing and TS-miRNA replacement systems. Modified anti-miRNA systems (microRNA mimetics–miRNA-mimic) can imitate or inhibit cellular miRNA functions to silence complex signal transduction pathways and control multiple physiologic functions [170]. Currently, miRNAs-based therapeutic approach constitutes a high-potential weapon to treat such heterogeneous disease, in terms of molecular abnormalities, pathologic tumor growth patterns and patients outcomes [171] as prostate cancer.
Currently, miRNAs are a part of multiple clinical trials exploring genomic signatures to solve a problem with patients’ selection and recognize companion biomarkers for conventional therapeutic approach. One of the prostate cancer clinical trial is interventional study for 50 patients with low-risk prostate cancer [tumor stage cT1-2a cN0 cM0 and Gleason Score of index lesion ≤ 6 (3 + 3)] to investigate the feasibility and toxicity of focal brachytherapy. A secondary outcome for this phase II clinical trial was to verify the correlation level of miR-141 and miR-375 expressions with visible efficacy of radiation intervention (ClinicalTrials.gov Identifier: NCT02391051). Previously, miR-141 together with miR-21 and miR-221 have been studied in the plasma of 51 prostate cancer patients with locally advanced and metastatic prostate cancer, where miR-21 was related to malignancy state and miR-141 was found to be capable of distinction localized and metastatic stage of prostate cancer [160]. Another one pre-clinical study of 667 samples of serum obtained from prostate cancer patients recognized miR-141 and miR-375 as molecules strongly involved in disease progression and were considered as non-invasive blood-based circulating biomarkers for prostate cancer progression [172]. More precisely, Brase and colleagues noticed that overexpression of miR-141, miR-200b and miR-375 in prostate cancer patients serum was associated with higher tumor stage and Gleason score [172]. Cheng et al. have analyzed panel of 365 miRNAs in prostate cancer serum and emerged miR-141, miR-200a, miR-200c, miR-375 and miR-210 molecules, which was significantly expressed in metastatic CRPC patients compared to age-matched healthy controls as well as in lymph metastasis samples [173]. In another study, Kelly et al. demonstrated a normalization in miR-141 expression in prostate cancer patients who underwent a radical retropubic-prostatectomy 10 days post-operation [174, 175]. Hao et al. have found miR-141 and miR-21 twofold up regulated, while miR-10, -16, -34c and -125b down regulated in FFPE tissue compared to BPH specimens [176]. Additionally, association of PSA level with miR-141 and miR-21 expressions enhanced the positive predictive value from 40 to 87.5% [177].
Improvement in survival rate for prostate cancer patients follows from the advances in first line treatment approach like surgery, radiation and antagonists to the androgen signaling axis [156]. However, the metastatic stage of prostate cancer has no treatment opportunities due to the resistance to androgen antagonists. In turn, FDA was approved taxane-based therapy to provide patients three months-longer overall survival [178, 179]. Another interventional study concern 211 prostate cancer patients at newly diagnosed metastatic stage (stage IV) (ClinicalTrials.gov Identifier: NCT01120236), were as a secondary outcome was measured correlation level between miR-141, miR-210, miR-200b and miR-375 as well as PSA level and circulating tumor cells (CTC). In this phase II randomized trial prostate cancer patients were treated with bicalutamide, goserelin, or leuprolide acetate in combination with cixutumumab or without cixutumumab. Another one observational cohort trial has recruiting status and attempts to identify the role of exosomal microRNA and define microRNA expression profiles to predict the aggressiveness of PCa in urine (ClinicalTrials.gov Identifier: NCT03911999).
Conclusion
PCa is one of the commonest malignancies in Western countries. The GLOBOCAN STATISTICS 2018 indicates at 7.1% of all new cancer incidence worldwide in 2018 year (for both sexes) [180]. MiRNA molecules offer new possibilities for diagnostic and systemic therapeutic approaches. They can attenuate or obstruct multiple networks involved in the tumor microenvironment, not only single transduction pathway [181]. Furthermore, miRNAs can be a more effective weapon against tumors as they are secreted from malignant cells and their anomalies in expression levels are associated with progression of prostate diseases.
We consider that the higher prognosis value is presented by sub-analysis associating miRNAs expression and GS > 6, showing association with miR-410-3p which was also connected with the presence of general metastasis and the pT stage, miR-503 with significance in general or lymph node metastasis and pT stage and miR-132 connected to general metastasis and distant metastasis. Mentioned miRNAs molecules could be used as a miRNA panel to determine the progression risk at early stages of the disease. Our findings present future potential therapeutic options to treat men with PCa and could provide better post-treatment care. The above mentioned experiments strongly indicated that miRNAs expression modification utilizing miRNA mimics or antagomirs can harmonize gene regulatory circuits and signal transduction pathways. Also, such activities may stop the aggressive status of tumors and reverse the cancerous status of cells.
Acknowledgements
The authors would like to thank their colleagues Joanna Ligmanowska for her assistance during tables creation and Michal Wlodarski for his valuable commentaries during the manuscript revision and support in language corrections of the manuscript.
Abbreviations
- PCa
Prostate cancer
- EMT
Epithelial–mesenchymal transition
- miRNAs
MicroRNAs
- mRNAs
Messenger RNAs
- Onc
Oncomirs
- FFPE
Formalin-fixed paraffin embedded tissue
- TNM
TNM Classification of Malignant Tumor System
- GS
Gleason score
- HRs
Hazard ratios
- RRs
Relative risks
- CIs
Confidence intervals
- I2
Inconsistency index
- NOS
Newcastle–Ottawa Scale
- RT-PCR
Real time polymerase chain reaction
- pT stage
Pathologic tumor stage
- TRAF5
Tumor necrosis factor receptor-associated factor 5
- CRC
Colorectal cancer
- MAPK
MAP kinase
- GC
Gastric cancer
- HIF-1α
Hypoxia-inducible factor
- PTEN
Phosphatase and tensin homolog deleted on chromosome ten
- FGF2
Fibroblast growth factor 2
- MEK1
MAPK kinase 1
- RKIP
Raf kinase inhibitor protein
Authors’ contributions
This paper was planned by MP, and she is responsible for the overall conception of the work, collecting information, data extraction process, methodology and initial drafting of the manuscript. AG were responsible for statistical elaboration. MP and AG were responsible for creating tables with important data as well as the graphic layout. MP and AG contributed critical revisions of the manuscript for the intellectual content, edited the manuscript and made the necessary language corrections. MB and DG supervised the work. All authors read and approved the final manuscript.
Funding
The authors received no specific funding for this work.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Martyna Parol, Email: mparol@cm.umk.pl.
Arkadiusz Gzil, Email: arkadiuszgzilresearch@gmail.com.
Magdalena Bodnar, Email: magdalena.bodnar@cm.umk.pl.
Dariusz Grzanka, Email: d_grzanka@cm.umk.pl.
References
- 1.Rawla P. Epidemiology of prostate cancer. Pathol Epidemiol Cancer. 2019;10(2):107–125. doi: 10.14740/wjon1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.American Cancer Society . Facts & figures 2019. Atlanta: American Cancer Society; 2019. [Google Scholar]
- 3.National Cancer Institute. Cancer stat facts: cancer of any site. Bethesda: National Cancer Institute; 2018.
- 4.Petrovich Z, Baert L, Bagshaw MA, Brady LW, Elgamal A, Goethuys H, et al. Adenocarcinoma of the prostate: Innovations in management. Am J Clin Oncol Cancer Clin Trials. 1997;20:111–119. doi: 10.1097/00000421-199704000-00001. [DOI] [PubMed] [Google Scholar]
- 5.Schulman KL, Kohles J. Economic burden of metastatic bone disease in the U.S. Cancer. 2007;109:2334–2342. doi: 10.1002/cncr.22678. [DOI] [PubMed] [Google Scholar]
- 6.Weiner AB, Matulewicz RS, Eggener SE, Schaeffer EM. Increasing incidence of metastatic prostate cancer in the United States (2004–2013) Prostate Cancer Prostatic Dis. 2016;19:395–397. doi: 10.1038/pcan.2016.30. [DOI] [PubMed] [Google Scholar]
- 7.Fidler IJ. The pathogenesis of cancer metastasis: the “seed and soil” hypothesis revisited. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 8.Thiery JP. Epithelial–mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 9.Zaravinos A. The regulatory role of MicroRNAs in EMT and cancer. J Oncol. 2015;2015:865816. doi: 10.1155/2015/865816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial–mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 11.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. Journal of Clinical Investigation. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 13.Esquela-Kerscher A, Slack FJ. Oncomirs—MicroRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 14.López-Camarillo C, Marchat LA. MicroRNAs in cancer; 2013. p. 1–426.
- 15.Shrestha S, Da HS, Huang WY, Huang HY, Chen W, Weng SL, et al. A systematic review of microRNA expression profiling studies in human gastric cancer. Cancer Med. 2014;3:878–888. doi: 10.1002/cam4.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 17.Dang TM, Wong WC, Ong SM, Li P, Lum J, Chen J, et al. MicroRNA expression profiling of human blood monocyte subsets highlights functional differences. Immunology. 2015;145:404–416. doi: 10.1111/imm.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 19.Zaravinos A, Lambrou GI, Mourmouras N, Katafygiotis P, Papagregoriou G, Giannikou K, et al. New miRNA profiles accurately distinguish renal cell carcinomas and upper tract urothelial carcinomas from the normal kidney. PLoS ONE. 2014;9:e91646. doi: 10.1371/journal.pone.0091646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wells GA, Shea B, O’connell D, Peterson J, Welch V, Losos M, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. Ottawa Hosp Res Inst; 2014 http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp. Accessed 13 Aug 2014.
- 21.Zhang Y, Zhang D, Lv J, Wang S, Zhang Q. miR-410–3p promotes prostate cancer progression via regulating PTEN/AKT/mTOR signaling pathway. Biochem Biophys Res Commun. 2018;503:2459–2465. doi: 10.1016/j.bbrc.2018.06.176. [DOI] [PubMed] [Google Scholar]
- 22.Wang L, Song G, Tan W, Qi M, Zhang L, Chan J, et al. miR-573 inhibits prostate cancer metastasis by regulating epithelial-mesenchymal transition. Oncotarget. 2015;6:35978–35990. doi: 10.18632/oncotarget.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bucay N, Bhagirath D, Sekhon K, Yang T, Fukuhara S, Majid S, et al. A novel microRNA regulator of prostate cancer epithelial-mesenchymal transition. Cell Death Differ. 2017;24:1263–1274. doi: 10.1038/cdd.2017.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nam RK, Benatar T, Wallis CJD, Amemiya Y, Yang W, Garbens A, et al. MiR-301a regulates E-cadherin expression and is predictive of prostate cancer recurrence. Prostate. 2016;76:869–884. doi: 10.1002/pros.23177. [DOI] [PubMed] [Google Scholar]
- 25.Colden M, Dar AA, Saini S, Dahiya PV, Shahryari V, Yamamura S, et al. MicroRNA-466 inhibits tumor growth and bone metastasis in prostate cancer by direct regulation of osteogenic transcription factor RUNX2. Cell Death Dis. 2017;8:e2572. doi: 10.1038/cddis.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang X, Chen Y, Du E, Yang K, Zhang Z, Qi S, et al. GATA3-driven expression of miR-503 inhibits prostate cancer progression by repressing ZNF217 expression. Cell Signal. 2016;28:1216–1224. doi: 10.1016/j.cellsig.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 27.Saini S, Majid S, Yamamura S, Tabatabai L, Suh SO, Shahryari V, et al. Regulatory role of mir-203 in prostate cancer progression and metastasis. Clin Cancer Res. 2011;17:5287–5298. doi: 10.1158/1078-0432.CCR-10-2619. [DOI] [PubMed] [Google Scholar]
- 28.Ren D, Yang Q, Dai Y, Guo W, Du H, Song L, et al. Oncogenic miR-210-3p promotes prostate cancer cell EMT and bone metastasis via NF-ΚB signaling pathway. Mol Cancer. 2017;16:117. doi: 10.1186/s12943-017-0688-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang D, Lu G, Shao Y, Xu D. microRNA-802 inhibits epithelial-mesenchymal transition through targeting flotillin-2 in human prostate cancer. Biosci Rep. 2017;37:BSR2016021. doi: 10.1042/BSR20160521. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Du Y, Liu XH, Zhu HC, Wang L, Ning JZ, Xiao CC. MiR-543 promotes proliferation and epithelial-mesenchymal transition in prostate cancer via targeting RKIP. Cell Physiol Biochem. 2017;41:1135–1146. doi: 10.1159/000464120. [DOI] [PubMed] [Google Scholar]
- 31.Fu W, Tao T, Qi M, Wang L, Hu J, Li X, et al. MicroRNA-132/212 upregulation inhibits TGF-β-mediated epithelial-mesenchymal transition of prostate cancer cells by targeting SOX4. Prostate. 2016;76(16):1560–1570. doi: 10.1002/pros.23241. [DOI] [PubMed] [Google Scholar]
- 32.Josson S, Gururajan M, Hu P, Shao C, Chu GCY, Zhau HE, et al. miR-409-3p/-5p promotes tumorigenesis, epithelial-to-mesenchymal transition, and bone metastasis of human prostate cancer. Clin Cancer Res. 2014;20:4636–4646. doi: 10.1158/1078-0432.CCR-14-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang S, Wa Q, Pan J, Peng X, Ren D, Huang Y, et al. Downregulation of miR-141-3p promotes bone metastasis via activating NF-κB signaling in prostate cancer. J Exp Clin Cancer Res. 2017;36:173. doi: 10.1186/s13046-017-0645-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guarnieri DJ, Dileone RJ. MicroRNAs: a new class of gene regulators. Ann Med. 2008;40:197–208. doi: 10.1080/07853890701771823. [DOI] [PubMed] [Google Scholar]
- 36.Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microRNAs: A review. J Physiol Biochem. 2011;67:129–139. doi: 10.1007/s13105-010-0050-6. [DOI] [PubMed] [Google Scholar]
- 37.Manikandan M, Deva Magendhra Rao AK, Arunkumar G, Manickavasagam M, Rajkumar KS, Rajaraman R, et al. Oral squamous cell carcinoma: MicroRNA expression profiling and integrative analyses for elucidation of tumourigenesis mechanism. Mol Cancer. 2016;15:28. doi: 10.1186/s12943-016-0512-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang W, Tian Y, Jiang S, Liu S, Zhao X, Tian D. MicroRNA-376c suppresses non-small-cell lung cancer cell growth and invasion by targeting LRH-1-mediated Wnt signaling pathway. Biochem Biophys Res Commun. 2016;473:980–986. doi: 10.1016/j.bbrc.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Wang RJ, Li JW, Bao BH, Wu HC, Du ZH, Su JL, et al. MicroRNA-873 (MiRNA-873) inhibits glioblastoma tumorigenesis and metastasis by suppressing the expression of IGF2BP1. J Biol Chem. 2015;290:8938–8948. doi: 10.1074/jbc.M114.624700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renjie W, Haiqian L. MiR-132, miR-15a and miR-16 synergistically inhibit pituitary tumor cell proliferation, invasion and migration by targeting Sox5. Cancer Lett. 2015;356:568–578. doi: 10.1016/j.canlet.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 41.Bucay N, Sekhon K, Majid S, Yamamura S, Shahryari V, Tabatabai ZL, et al. Novel tumor suppressor microRNA at frequently deleted chromosomal region 8p21 regulates epidermal growth factor receptor in prostate cancer. Oncotarget. 2016;7:70388–70403. doi: 10.18632/oncotarget.11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu M, Wang T, He M, Cheng W, Yan T, Huang Z, et al. Tumor suppressor role of miR-3622b-5p in ERBB2-positive cancer. Oncotarget. 2017;8:23008–23019. doi: 10.18632/oncotarget.14968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu S, Luan T, Jiang C, Huang Y, Li N, Wang H, et al. miR-3622a promotes proliferation and invasion of bladder cancer cells by downregulating LASS2. Gene. 2019;701:23–31. doi: 10.1016/j.gene.2019.02.083. [DOI] [PubMed] [Google Scholar]
- 44.Chang S, Sun G, Zhang D, Li Q, Qian H. MiR-3622a-3p acts as a tumor suppressor in colorectal cancer by reducing stemness features and EMT through targeting spalt-like transcription factor 4. Cell Death Dis. 2020;11:592. doi: 10.1038/s41419-020-02789-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Onodera Y, Miki Y, Suzuki T, Takagi K, Akahira JI, Sakyu T, et al. Runx2 in human breast carcinoma: Its potential roles in cancer progression. Cancer Sci. 2010;101:2670–2675. doi: 10.1111/j.1349-7006.2010.01742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tong F, Ying Y, Pan H, Zhao W, Li H, Zhan X. Microrna-466 (Mir-466) functions as a tumor suppressor and prognostic factor in colorectal cancer (CRC) Bosn J Basic Med Sci. 2018;18:252–259. doi: 10.17305/bjbms.2018.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun J, Zhou J, Dong M, Sheng W. Dysregulation of MicroRNA-543 expression in colorectal cancer promotes tumor migration and invasion. Mol Carcinog. 2017;56:250–257. doi: 10.1002/mc.22489. [DOI] [PubMed] [Google Scholar]
- 48.Yu L, Zhou L, Cheng Y, Sun L, Fan J, Liang J, et al. MicroRNA-543 acts as an oncogene by targeting PAQR3 in hepatocellular carcinoma. Am J Cancer Res. 2014;4:897–906. [PMC free article] [PubMed] [Google Scholar]
- 49.Li J, Dong G, Wang B, Gao W, Yang Q. MiR-543 promotes gastric cancer cell proliferation by targeting SIRT1. Biochem Biophys Res Commun. 2016;469:15–21. doi: 10.1016/j.bbrc.2015.11.062. [DOI] [PubMed] [Google Scholar]
- 50.Bi M, Chen W, Yu H, Wang J, Ding F, Tang DJ, et al. miR-543 is up-regulated in gefitinib-resistant non-small cell lung cancer and promotes cell proliferation and invasion via phosphatase and tensin homolog. Biochem Biophys Res Commun. 2016;480:369–374. doi: 10.1016/j.bbrc.2016.10.055. [DOI] [PubMed] [Google Scholar]
- 51.Liu G, Zhou JP, Dong M. Down-regulation of miR-543 expression increases the sensitivity of colorectal cancer cells to 5-Fluorouracil through the PTEN/PI3K/AKT pathway. Biosci Rep. 2019;39:BSR20190249. doi: 10.1042/BSR20190249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu L, Yu J, Wang Z, Zhu Q, Wang W, Lan Q. MiR-543 functions as a tumor suppressor in glioma in vitro and in vivo. Oncol Rep. 2017;38:725–734. doi: 10.3892/or.2017.5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li B, Chen H, Shang LX, Gao W. MicroRNA-543 suppresses endometrial cancer oncogenicity via targeting FAK and TWIST1 expression. Arch Gynecol Obstet. 2014;290:533–541. doi: 10.1007/s00404-014-3219-3. [DOI] [PubMed] [Google Scholar]
- 54.Zhao JL, Zhang L, Guo X, Wang JH, Zhou W, Liu M, et al. MiR-212/132 downregulates SMAD2 expression to suppress the G1/S phase transition of the cell cycle and the epithelial to mesenchymal transition in cervical cancer cells. IUBMB Life. 2015;67:380–394. doi: 10.1002/iub.1381. [DOI] [PubMed] [Google Scholar]
- 55.Lei CJ, Li L, Gao X, Zhang J, Pan QY, Long HC, et al. Hsa-miR-132 inhibits proliferation of hepatic carcinoma cells by targeting YAP. Cell Biochem Funct. 2015;33:326–333. doi: 10.1002/cbf.3119. [DOI] [PubMed] [Google Scholar]
- 56.You J, Li Y, Fang N, Liu B, Zu L, Chang R, et al. MiR-132 suppresses the migration and invasion of lung cancer cells via targeting the EMT regulator ZEB2. PLoS ONE. 2014;9:e91827. doi: 10.1371/journal.pone.0091827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang B, Lu L, Zhang X, Ye W, Wu J, Xi Q, et al. Hsa-miR-132 regulates apoptosis in non-small cell lung cancer independent of acetylcholinesterase. J Mol Neurosci. 2014;53:335–344. doi: 10.1007/s12031-013-0136-z. [DOI] [PubMed] [Google Scholar]
- 58.Liu K, Li X, Cao Y, Ge Y, Wang J, Shi B. MiR-132 inhibits cell proliferation, invasion and migration of hepatocellular carcinoma by targeting PIK3R3. Int J Oncol. 2015;47:1585–1593. doi: 10.3892/ijo.2015.3112. [DOI] [PubMed] [Google Scholar]
- 59.Wanet A, Tacheny A, Arnould T, Renard P. MiR-212/132 expression and functions: Within and beyond the neuronal compartment. Nucleic Acids Res. 2012;40:4742–4753. doi: 10.1093/nar/gks151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Medina PP, Slack FJ. microRNAs and cancer: an overview. Cell Cycle. 2008;7:2485–2492. doi: 10.4161/cc.7.16.6453. [DOI] [PubMed] [Google Scholar]
- 61.Chen JQ, Ou YL, Huang ZP, Hong YG, Tao YP, Wang ZG, et al. MicroRNA-212-3p inhibits the proliferation and invasion of human hepatocellular carcinoma cells by suppressing CTGF expression. Sci Rep. 2019;9:9820. doi: 10.1038/s41598-019-46088-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lv ZD, Yang DX, Liu XP, Jin LY, Wang XG, Yang ZC, et al. MiR-212-5p suppresses the epithelial-mesenchymal transition in triple-negative breast cancer by targeting Prrx2. Cell Physiol Biochem. 2018;44:1785–1795. doi: 10.1159/000485785. [DOI] [PubMed] [Google Scholar]
- 63.Tili E, Michaille JJ, Piurowski V, Rigot B, Croce CM. MicroRNAs in intestinal barrier function, inflammatory bowel disease and related cancers—their effects and therapeutic potentials. Curr Opin Pharmacol. 2017;37:142–150. doi: 10.1016/j.coph.2017.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 65.Gao Y, Feng B, Han S, Zhang K, Chen J, Li C, et al. The roles of MicroRNA-141 in human cancers: from diagnosis to treatment. Cell Physiol Biochem. 2016;38:427–448. doi: 10.1159/000438641. [DOI] [PubMed] [Google Scholar]
- 66.Fang M, Huang W, Wu X, Gao Y, Ou J, Zhang X, et al. MiR-141-3p Suppresses tumor growth and metastasis in papillary thyroid cancer via targeting Yin Yang 1. Anat Rec. 2019;302:258–268. doi: 10.1002/ar.23940. [DOI] [PubMed] [Google Scholar]
- 67.Zhou Y, Zhong JH, Gong FS, Xiao J. MiR-141-3p suppresses gastric cancer induced transition of normal fibroblast and BMSC to cancer-associated fibroblasts via targeting STAT4. Exp Mol Pathol. 2019;107:85–94. doi: 10.1016/j.yexmp.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 68.Li W, Cui Y, Wang D, Wang Y, Wang L. MiR-141-3p functions as a tumor suppressor through directly targeting ZFR in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;509(3):647–656. doi: 10.1016/j.bbrc.2018.12.089. [DOI] [PubMed] [Google Scholar]
- 69.Liu CZ, Ye ZH, Ma J, He RQ, Liang HW, Peng ZG, et al. A qRT-PCR and gene functional enrichment study focused on downregulation of miR-141-3p in hepatocellular carcinoma and its clinicopathological significance. Technol Cancer Res Treat. 2017;16:835–849. doi: 10.1177/1533034617705056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hui L, Bakiri L, Mairhorfer A, Schweifer N, Haslinger C, Kenner L, et al. p38α suppresses normal and cancer cell proliferation by antagonizing the JNK-c-Jun pathway. Nat Genet. 2007;39:741–749. doi: 10.1038/ng2033. [DOI] [PubMed] [Google Scholar]
- 71.Long J, Ou C, Xia H, Zhu Y, Liu D. MiR-503 inhibited cell proliferation of human breast cancer cells by suppressing CCND1 expression. Tumor Biol. 2015;36:8697–8702. doi: 10.1007/s13277-015-3623-8. [DOI] [PubMed] [Google Scholar]
- 72.Zhou B, Ma R, Si W, Li S, Xu Y, Tu X, et al. MicroRNA-503 targets FGF2 and VEGFA and inhibits tumor angiogenesis and growth. Cancer Lett. 2013;333:159–169. doi: 10.1016/j.canlet.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 73.Peng Y, Liu YM, Li LC, Wang LL, Wu XL. MicroRNA-503 inhibits gastric cancer cell growth and epithelial-to-mesenchymal transition. Oncol Lett. 2014;7:1233–1238. doi: 10.3892/ol.2014.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guo J, Liu X, Wang M. MiR-503 suppresses tumor cell proliferation and metastasis by directly targeting RNF31 in prostate cancer. Biochem Biophys Res Commun. 2015;464:1302–1308. doi: 10.1016/j.bbrc.2015.07.127. [DOI] [PubMed] [Google Scholar]
- 75.Yin ZL, Wang YL, Ge SF, Guo TT, Wang L, Zheng XM, et al. Reduced expression of miR-503 is associated with poor prognosis in cervical cancer. Eur Rev Med Pharmacol Sci. 2015;19:4081–4085. [PubMed] [Google Scholar]
- 76.Li B, Liu L, Li X, Wu L. MiR-503 suppresses metastasis of hepatocellular carcinoma cell by targeting PRMT1. Biochem Biophys Res Commun. 2015;464:982–987. doi: 10.1016/j.bbrc.2015.06.169. [DOI] [PubMed] [Google Scholar]
- 77.Chang SW, Yue J, Wang BC, Zhang XL. MiR-503 inhibits cell proliferation and induces apoptosis in colorectal cancer cells by targeting E2F3. Int J Clin Exp Pathol. 2015;8:12853–12860. [PMC free article] [PubMed] [Google Scholar]
- 78.Xu X, Chen H, Lin Y, Hu Z, Mao Y, Wu J, et al. Microrna-409-3p inhibits migration and invasion of bladder cancer cells via targeting c-met. Mol Cells. 2013;36:62–68. doi: 10.1007/s10059-013-0044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li C, Nie H, Wang M, Su L, Li J, Yu B, et al. MicroRNA-409-3p regulates cell proliferation and apoptosis by targeting PHF10 in gastric cancer. Cancer Lett. 2012;320:189–197. doi: 10.1016/j.canlet.2012.02.030. [DOI] [PubMed] [Google Scholar]
- 80.Weng C, Dong H, Chen G, Zhai Y, Bai R, Hu H, et al. MiR-409-3p inhibits HT1080 cell proliferation, vascularization and metastasis by targeting angiogenin. Cancer Lett. 2012;323:171–179. doi: 10.1016/j.canlet.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 81.Josson S, Gururajan M, Sung SY, Hu P, Shao C, Zhau HE, et al. Stromal fibroblast-derived miR-409 promotes epithelial-to-mesenchymal transition and prostate tumorigenesis. Oncogene. 2015;34:2690–2699. doi: 10.1038/onc.2014.212. [DOI] [PubMed] [Google Scholar]
- 82.Zheng JZ, Huang YN, Yao L, Liu YR, Liu S, Hu X, et al. Elevated miR-301a expression indicates a poor prognosis for breast cancer patients. Sci Rep. 2018;8:2225. doi: 10.1038/s41598-018-20680-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li X, Zhong M, Wang J, Wang L, Lin Z, Cao Z, et al. MiR-301a promotes lung tumorigenesis by suppressing Runx3. Mol Cancer. 2019;18:99. doi: 10.1186/s12943-019-1024-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang K-D, Hu B, Cen G, Yang Y-H, Chen W-W, Guo Z-Y, et al. MiR-301a transcriptionally activated by HIF-2α promotes hypoxia-induced epithelial-mesenchymal transition by targeting TP63 in pancreatic cancer. World J Gastroenterol. 2020;26:2349–2373. doi: 10.3748/wjg.v26.i19.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang W, Qu A, Liu W, Liu Y, Zheng G, Du L, et al. Circulating miR-210 as a diagnostic and prognostic biomarker for colorectal cancer. Eur J Cancer Care (Engl) 2017;26:e12448. doi: 10.1111/ecc.12448. [DOI] [PubMed] [Google Scholar]
- 86.Qin Q, Wei F, Li B. Multiple functions of hypoxia-regulated miR-210 in cancer. J Exp Clin Cancer Res. 2014;33:50. doi: 10.1186/1756-9966-33-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang J, Zhao J, Shi M, Ding Y, Sun H, Yuan F, et al. Elevated expression of miR-210 predicts poor survival of cancer patients: a systematic review and meta-analysis. PLoS ONE. 2014;9:e89223. doi: 10.1371/journal.pone.0089223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bavelloni A, Ramazzotti G, Poli A, Piazzi M, Focaccia E, Blalock W, et al. Mirna-210: a current overview. Anticancer Res. 2017;37:6511–6521. doi: 10.21873/anticanres.12107. [DOI] [PubMed] [Google Scholar]
- 89.Jung KO, Youn H, Lee CH, Kang KW, Chung JK. Visualization of exosome-mediated miR-210 transfer from hypoxic tumor cells. Oncotarget. 2017;8:9899–9910. doi: 10.18632/oncotarget.14247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Viticchiè G, Lena AM, Latina A, Formosa A, Gregersen LH, Lund AH, et al. MiR-203 controls proliferation, migration and invasive potential of prostate cancer cell lines. Cell Cycle. 2011;10:1121–1131. doi: 10.4161/cc.10.7.15180. [DOI] [PubMed] [Google Scholar]
- 91.Boll K, Reiche K, Kasack K, Mörbt N, Kretzschmar AK, Tomm JM, et al. MiR-130a, miR-203 and miR-205 jointly repress key oncogenic pathways and are downregulated in prostate carcinoma. Oncogene. 2013;32:277–278. doi: 10.1038/onc.2012.55. [DOI] [PubMed] [Google Scholar]
- 92.Wang B, Li X, Zhao G, Yan H, Dong P, Watari H, et al. miR-203 inhibits ovarian tumor metastasis by targeting BIRC5 and attenuating the TGFβ pathway. J Exp Clin Cancer Res. 2018;37:235. doi: 10.1186/s13046-018-0906-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ting W, Feng C, Zhang M, Long F, Bai M. Overexpression of microRNA-203 suppresses proliferation, invasion, and migration while accelerating apoptosis of CSCC cell line SCL-1. Mol Ther Nucleic Acids. 2020;21:428–440. doi: 10.1016/j.omtn.2020.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 94.Hu YW, Chen ZP, Hu XM, Zhao JY, Huang JL, Ma X, et al. The miR-573/apoM/Bcl2A1-dependent signal transduction pathway is essential for hepatocyte apoptosis and hepatocarcinogenesis. Apoptosis. 2015;20:1321–1337. doi: 10.1007/s10495-015-1153-x. [DOI] [PubMed] [Google Scholar]
- 95.Danza K, De SS, Pinto R, Pilato B, Palumbo O, Merla G, et al. MiR-578 and miR-573 as potential players in BRCA-related breast cancer angiogenesis. Oncotarget. 2015;6:471–483. doi: 10.18632/oncotarget.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang L, Song G, Tan W, Qi M, Zhang L, Chan J, et al. miR-573 inhibits prostate cancer metastasis by regulating epithelialmesenchymal transition. Oncotarget. 2015;6:35978–35990. doi: 10.18632/oncotarget.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang Y, Cao J, Zhang S, Sun L, Nan Y, Yao H, et al. MicroRNA-802 induces hepatitis B virus replication and replication through regulating SMARCE1 expression in hepatocellular carcinoma. Cell Death Dis. 2019;10:783. doi: 10.1038/s41419-019-1999-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang B, Sun L, Liang L. MiRNA-802 suppresses proliferation and migration of epithelial ovarian cancer cells by targeting YWHAZ. J Ovarian Res. 2019;12:100. doi: 10.1186/s13048-019-0576-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sansom SE, Nuovo GJ, Martin MM, Kotha SR, Parinandi NL, Elton TS. miR-802 regulates human angiotensin II type 1 receptor expression in intestinal epithelial C2BBe1 cells. Am J Physiol Gastrointest Liver Physiol. 2010;299(3):G632–G642. doi: 10.1152/ajpgi.00120.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang XY, Mu JH, Liu LY, Zhang HZ. Upregulation of miR-802 suppresses gastric cancer oncogenicity via targeting RAB23 expression. Eur Rev Med Pharmacol Sci. 2017;21:4071–4078. [PubMed] [Google Scholar]
- 101.Grzywa TM, Klicka K, Rak B, Mehlich D, Garbicz F, Zieliński G, et al. Lineage-dependent role of miR-410-3p as oncomiR in gonadotroph and corticotroph pituitary adenomas or tumor suppressor miR in somatotroph adenomas via MAPK, PTEN/AKT, and STAT3 signaling pathways. Endocrine. 2019;65:646–655. doi: 10.1007/s12020-019-01960-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wen R, Umeano AC, Essegian DJ, Sabitaliyevich UY, Wang K, Farooqi AA. Role of microRNA-410 in molecular oncology: a double edged sword. J Cell Biochem. 2018;119:8737–8742. doi: 10.1002/jcb.27251. [DOI] [PubMed] [Google Scholar]
- 103.Xiong J, Wang D, Wei A, Ke N, Wang Y, Tang J, et al. MicroRNA-410-3p attenuates gemcitabine resistance in pancreatic ductal adenocarcinoma by inhibiting HMGB1-mediated autophagy. Oncotarget. 2017;8:107500–107512. doi: 10.18632/oncotarget.22494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Abramovic I, Ulamec M, Bojanac Katusic A, Bulic-Jakus F, Jezek D, Sincic N. MiRNA in prostate cancer: challenges toward translation. Epigenomics. 2020;12:543–558. doi: 10.2217/epi-2019-0275. [DOI] [PubMed] [Google Scholar]
- 105.Zhou H, Zhu X. MicroRNA-21 and microRNA-30C as diagnostic biomarkers for prostate cancer: a meta-analysis. Cancer Manag Res. 2019;11:2039–2050. doi: 10.2147/CMAR.S189026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Song CJ, Chen H, Chen LZ, Ru GM, Guo JJ, Ding QN. The potential of microRNAs as human prostate cancer biomarkers: a meta-analysis of related studies. J Cell Biochem. 2018;119:2763–2786. doi: 10.1002/jcb.26445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zedan AH, Osther PJS, Assenholt J, Madsen JS, Hansen TF. Circulating miR-141 and miR-375 are associated with treatment outcome in metastatic castration resistant prostate cancer. Sci Rep. 2020;10:227. doi: 10.1038/s41598-019-57101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pashaei E, Pashaei E, Ahmady M, Ozen M, Aydin N. Meta-analysis of miRNA expression profiles for prostate cancer recurrence following radical prostatectomy. PLoS ONE. 2017;12:e0179543. doi: 10.1371/journal.pone.0179543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Guan X. Cancer metastases: Challenges and opportunities. Acta Pharm Sin B. 2015;5:402–418. doi: 10.1016/j.apsb.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, et al. Hsa-miR-210 is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res. 2008;14:1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 111.Puisségur MP, Mazure NM, Bertero T, Pradelli L, Grosso S, Robbe-Sermesant K, et al. MiR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 2011;18:465–478. doi: 10.1038/cdd.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Greither T, Grochola LF, Udelnow A, Lautenschläger C, Würl P, Taubert H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer. 2010;126:73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 113.Gee HE, Camps C, Buffa FM, Patiar S, Winter SC, Betts G, et al. hsa-mir-210 is a marker of tumor hypoxia and a prognostic factor in head and neck cancer. Cancer. 2010;116:2148–2158. doi: 10.1002/cncr.25009. [DOI] [PubMed] [Google Scholar]
- 114.Pasculli B, Barbano R, Rendina M, Fontana A, Copetti M, Mazza T, et al. Hsa-miR-210-3p expression in breast cancer and its putative association with worse outcome in patients treated with Docetaxel. Sci Rep. 2019;9(1):1–9. doi: 10.1038/s41598-019-51581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Foekens JA, Sieuwerts AM, Smid M, Look MP, De Weerd V, Boersma AWM, et al. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci U S A. 2008;105:13021–13026. doi: 10.1073/pnas.0803304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Volinia S, Shaw AS, Peddi PF, Galasso M, Kornev AP, Ellis MJ, et al. Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNA. Proc Natl Acad Sci U S A. 2012;109:3024–3029. doi: 10.1073/pnas.1200010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liang Z, Li X, Liu S, Li C, Wang X, Xing J. MiR-141–3p inhibits cell proliferation, migration and invasion by targeting TRAF5 in colorectal cancer. Biochem Biophys Res Commun. 2019;514(3):699–705. doi: 10.1016/j.bbrc.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 118.Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumor suppression. Cell. 2000;100:387–390. doi: 10.1016/s0092-8674(00)80674-1. [DOI] [PubMed] [Google Scholar]
- 119.Gruber Filbin M, Dabral SK, Pazyra-Murphy MF, Ramkissoon S, Kung AL, Pak E, et al. Coordinate activation of Shh and PI3K signaling in PTEN-deficient glioblastoma: new therapeutic opportunities. Nat Med. 2013;19:1518–1523. doi: 10.1038/nm.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Siveen KS, Ahn KS, Ong TH, Shanmugam MK, Li F, Yap WN, et al. γ-tocotrienol inhibits angiogenesis-dependent growth of human hepatocellular carcinoma through abrogation of AKT/mTOR pathway in an orthotopic mouse model. Oncotarget. 2014;5:1897–1911. doi: 10.18632/oncotarget.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yeung K, Seitz T, Li S, Janosch P, McFerran B, Kaiser C, et al. Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature. 1999;401:173–177. doi: 10.1038/43686. [DOI] [PubMed] [Google Scholar]
- 122.Yeung KC, Rose DW, Dhillon AS, Yaros D, Gustafsson M, Chatterjee D, et al. Raf kinase inhibitor protein interacts with NF-B-inducing kinase and TAK1 and Inhibits NF-B activation. Mol Cell Biol. 2001;21:7207–7217. doi: 10.1128/MCB.21.21.7207-7217.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Haga CL, Phinney DG. MicroRNAs in the imprinted DLK1-DIO3 region repress the epithelial-to-mesenchymal transition by targeting the TWIST1 protein signaling network. J Biol Chem. 2012;287:42695–42707. doi: 10.1074/jbc.M112.387761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wu J, Gao F, Xu T, Deng X, Wang C, Yang X, et al. MiR-503 suppresses the proliferation and metastasis of esophageal squamous cell carcinoma by triggering autophagy via PKA/mTOR signaling. Int J Oncol. 2018;52(5):1427–1442. doi: 10.3892/ijo.2018.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yang Y, Liu L, Zhang Y, Guan H, Wu J, Zhu X, et al. MiR-503 targets PI3K p85 and IKK-β and suppresses progression of non-small cell lung cancer. Int J Cancer. 2014;135:1531–1542. doi: 10.1002/ijc.28799. [DOI] [PubMed] [Google Scholar]
- 126.Qiu T, Zhou L, Wang T, Xu J, Wang J, Chen W, et al. MiR-503 regulates the resistance of non-small cell lung cancer cells to cisplatin by targeting Bcl-2. Int J Mol Med. 2013;32:593–598. doi: 10.3892/ijmm.2013.1439. [DOI] [PubMed] [Google Scholar]
- 127.Wang D, Ren J, Ren H, Fu JL, Yu D. MicroRNA-132 suppresses cell proliferation in human breast cancer by directly targeting FOXA1. Acta Pharmacol Sin. 2018;39:124–131. doi: 10.1038/aps.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wong TS, Liu XB, Wong BYH, Ng RWM, Yuen APW, Wei WI. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res. 2008;14:2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- 129.Chung YW, Bae HS, Song JY, Lee JK, Lee NW, Kim T, et al. Detection of microRNA as novel biomarkers of epithelial ovarian cancer from the serum of ovarian cancer patient. Int J Gynecol Cancer. 2013;23:673–679. doi: 10.1097/IGC.0b013e31828c166d. [DOI] [PubMed] [Google Scholar]
- 130.Formosa A, Lena AM, Markert EK, Cortelli S, Miano R, Mauriello A, et al. DNA methylation silences miR-132 in prostate cancer. Oncogene. 2013;32:127–134. doi: 10.1038/onc.2012.14. [DOI] [PubMed] [Google Scholar]
- 131.Humphrey PA. Gleason grading and prognostic factors in carcinoma of the prostate. Mod Pathol. 2004;17:292–306. doi: 10.1038/modpathol.3800054. [DOI] [PubMed] [Google Scholar]
- 132.Diagnosis and treatment of prostate cancer: Summary of NICE guidance. BMJ (Online). 2014. [DOI] [PMC free article] [PubMed]
- 133.Yue H, Liu L, Song Z. miR-212 regulated by HIF-1α promotes the progression of pancreatic cancer. Exp Ther Med. 2019;17:2359–2365. doi: 10.3892/etm.2019.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhou C, Tan DM, Chen L, Xu XY, Sun CC, Zong LJ, et al. Effect of miR-212 targeting TCF7L2 on the proliferation and metastasis of cervical cancer. Eur Rev Med Pharmacol Sci. 2017;21:219–226. [PubMed] [Google Scholar]
- 135.Jiang X, Chen X, Chen L, Ma Y, Zhou L, Qi Q, et al. Upregulation of the miR-212/132 cluster suppresses proliferation of human lung cancer cells. Oncol Rep. 2015;33:705–712. doi: 10.3892/or.2014.3637. [DOI] [PubMed] [Google Scholar]
- 136.Xie M, Fu Z, Cao J, Liu Y, Wu J, Li Q, et al. MicroRNA-132 and microRNA-212 mediate doxorubicin resistance by down-regulating the PTEN-AKT/NF-κB signaling pathway in breast cancer. Biomed Pharmacother. 2018;102:286–294. doi: 10.1016/j.biopha.2018.03.088. [DOI] [PubMed] [Google Scholar]
- 137.Wang HF, Chen H, Ma MW, Wang JA, Tang TT, Ni LS, et al. miR-573 regulates melanoma progression by targeting the melanoma cell adhesion molecule. Oncol Rep. 2013;30:520–526. doi: 10.3892/or.2013.2451. [DOI] [PubMed] [Google Scholar]
- 138.Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2009;31:766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 139.Muhammad N, Bhattacharya S, Steele R, Ray RB. Anti-miR-203 suppresses ER-positive breast cancer growth and stemness by targeting SOCS3. Oncotarget. 2016;7:58595. doi: 10.18632/oncotarget.11193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang C, Zheng X, Shen C, Shi Y. MicroRNA-203 suppresses cell proliferation and migration by targeting BIRC5 and LASP1 in human triple-negative breast cancer cells. J Exp Clin Cancer Res. 2012;31:58. doi: 10.1186/1756-9966-31-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhang F, Yang Z, Cao M, Xu Y, Li J, Chen X, et al. MiR-203 suppresses tumor growth and invasion and down-regulates MiR-21 expression through repressing Ran in esophageal cancer. Cancer Lett. 2014;342:121–129. doi: 10.1016/j.canlet.2013.08.037. [DOI] [PubMed] [Google Scholar]
- 142.He JH, Li YM, Li YG, Xie XY, Wang L, Chun SY, et al. hsa-miR-203 enhances the sensitivity of leukemia cells to arsenic trioxide. Exp Ther Med. 2013;5:1315–1321. doi: 10.3892/etm.2013.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yang CH, Wang Y, Sims M, Cai C, He P, Häcker H, et al. MicroRNA203a suppresses glioma tumorigenesis through an ATM-dependent interferon response pathway. Oncotarget. 2017;8:112980–112991. doi: 10.18632/oncotarget.22945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhang X, Zhang Y, Liu X, Fang A, Li P, Li Z, et al. MicroRNA-203 is a prognostic indicator in bladder cancer and enhances chemosensitivity to cisplatin via apoptosis by targeting Bcl-w and Survivin. PLoS ONE. 2015;10:e0143441. doi: 10.1371/journal.pone.0143441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Xiaohong Z, Lichun F, Na X, Kejian Z, Xiaolan X, Shaosheng W. MiR-203 promotes the growth and migration of ovarian cancer cells by enhancing glycolytic pathway. Tumor Biol. 2016;37:14989–14997. doi: 10.1007/s13277-016-5415-1. [DOI] [PubMed] [Google Scholar]
- 146.Zhao G, Wang Q, Gu Q, Qiang W, Wei JJ, Dong P, et al. Lentiviral CRISPR/Cas9 nickase vector mediated BIRC5 editing inhibits epithelial to mesenchymal transition in ovarian cancer cells. Oncotarget. 2017;8:94666–94680. doi: 10.18632/oncotarget.21863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chen LZ, Ding Z, Zhang Y, He ST, Wang XH. MiR-203 over-expression promotes prostate cancer cell apoptosis and reduces ADM resistance. Eur Rev Med Pharmacol Sci. 2018;22:3734–3741. doi: 10.26355/eurrev_201806_15253. [DOI] [PubMed] [Google Scholar]
- 148.Cosma G, Acampora G, Brown D, Rees RC, Khan M, Pockley AG. Prediction of pathological stage in patients with prostate cancer: a neuro-fuzzy model. PLoS ONE. 2016;11:e015585. doi: 10.1371/journal.pone.0155856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Du B, Wu D, Yang X, Wang T, Shi X, Lv Y, et al. The expression and significance of microRNA in different stages of colorectal cancer. Med (United States) 2018;97:e9635. doi: 10.1097/MD.0000000000009635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhang YF, Yu Y, Song WZ, Zhang RM, Jin S, Bai JW, et al. miR-410-3p suppresses breast cancer progression by targeting Snail. Oncol Rep. 2016;36:480–486. doi: 10.3892/or.2016.4828. [DOI] [PubMed] [Google Scholar]
- 151.Zhang L, Pang Y, Cui X, Jia W, Cui W, Liu Y, et al. Microrna-410-3p upregulation suppresses proliferation, invasion and migration, and promotes apoptosis in rhabdomyosarcoma cells. Oncol Lett. 2019;18:936–943. doi: 10.3892/ol.2019.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang W, He Y, Rui J, Xu MQ. Mir-410 acts as an oncogene in colorectal cancer cells by targeting dickkopf-related protein 1 via the Wnt/β-catenin signaling pathway. Oncol Lett. 2019;17(1):807–814. doi: 10.3892/ol.2018.9710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhao H, Diao C, Wang X, Xie Y, Liu Y, Gao X, et al. MiR-543 promotes migration, invasion and epithelial-mesenchymal transition of esophageal cancer cells by targeting phospholipase A2 group IVA. Cell Physiol Biochem. 2018;48:1595–1604. doi: 10.1159/000492281. [DOI] [PubMed] [Google Scholar]
- 154.Fan C, Lin Y, Mao Y, Huang Z, Liu AY, Ma H, et al. MicroRNA-543 suppresses colorectal cancer growth and metastasis by targeting KRAS, MTA1 and HMGA2. Oncotarget. 2016;7:21825–21839. doi: 10.18632/oncotarget.7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ke XS, Liu CM, Liu DP, Liang CC. MicroRNAs: Key participants in gene regulatory networks. Curr Opin Chem Biol. 2003;7:516–523. doi: 10.1016/s1367-5931(03)00075-9. [DOI] [PubMed] [Google Scholar]
- 156.Smith B, Agarwal P, Bhowmick NA. MicroRNA applications for prostate, ovarian and breast cancer in the era of precision medicine. Endocr Relat Cancer. 2017;24:R157–R172. doi: 10.1530/ERC-16-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Braicu C, Buiga R, Cojocneanu R, Buse M, Raduly L, Pop LA, et al. Connecting the dots between different networks: MiRNAs associated with bladder cancer risk and progression. J Exp Clin Cancer Res. 2019;38:433. doi: 10.1186/s13046-019-1406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Aghdam AM, Amiri A, Salarinia R, Masoudifar A, Ghasemi F, Mirzaei H. MicroRNAs as diagnostic, prognostic, and therapeutic biomarkers in prostate. Cancer. 2019;29(2):127–139. doi: 10.1615/CritRevEukaryotGeneExpr.2019025273. [DOI] [PubMed] [Google Scholar]
- 159.Watahiki Y, Macfarlane RJ, Gleave ME, Crea F, Wang Y, Helgason CD, et al. Plasma miRNAs as biomarkers to identify patients with castration-resistant metastatic prostate cancer. Int J Mol Sci. 2013;14:7757–7770. doi: 10.3390/ijms14047757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Agaoglu FY, Kovancilar M, Dizdar Y, Darendeliler E, Holdenrieder S, Dalay N, et al. Investigation of miR-21, miR-141, and miR-221 in blood circulation of patients with prostate cancer. Tumor Biol. 2011;32:583–588. doi: 10.1007/s13277-011-0154-9. [DOI] [PubMed] [Google Scholar]
- 161.Porzycki P, Ciszkowicz E, Semik M, Tyrka M. Combination of three miRNA (miR-141, miR-21, and miR-375) as potential diagnostic tool for prostate cancer recognition. Int Urol Nephrol. 2018;50:1619–1626. doi: 10.1007/s11255-018-1938-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kolluru V, Chandrasekaran B, Tyagi A, Dervishi A, Ankem M, Yan X, et al. miR-301a expression: diagnostic and prognostic marker for prostate cancer. Urol Oncol Semin Orig Investig. 2018;36:503. doi: 10.1016/j.urolonc.2018.07.014. [DOI] [PubMed] [Google Scholar]
- 163.Bertoli G, Cava C, Castiglioni I. MicroRNAs as biomarkers for diagnosis, prognosis and theranostics in prostate cancer. Int J Mol Sci. 2016;17:421. doi: 10.3390/ijms17030421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Loblaw DA, Walker-Dilks C, Winquist E, Hotte SJ. Systemic therapy in men with metastatic castration-resistant prostate cancer: a systematic review. Clin Oncol. 2013;32:3436. doi: 10.1016/j.clon.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 165.Zedan AH, Osther PJS, Assenholt J, Madsen JS, Hansen T. Association between circulating miRNA-141 and miRNA-375 and treatment outcome in metastatic castration-resistant prostate cancer. J Clin Oncol. 2019;37:e16507. [Google Scholar]
- 166.Gonzales JC, Fink LM, Goodman OB, Symanowski JT, Vogelzang NJ, Ward DC. Comparison of circulating MicroRNA 141 to circulating tumor cells, lactate dehydrogenase, and prostate-specific antigen for determining treatment response in patients with metastatic prostate cancer. Clin Genitourin Cancer. 2011;9:39–45. doi: 10.1016/j.clgc.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 167.Nguyen HCN, Xie W, Yang M, Hsieh CL, Drouin S, Lee GSM, et al. Expression differences of circulating microRNAs in metastatic castration resistant prostate cancer and low-risk, localized prostate cancer. Prostate. 2013;73:346–354. doi: 10.1002/pros.22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TLJ, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 169.Srivastava A, Goldberger H, Dimtchev A, Ramalinga M, Chijioke J, Marian C, et al. MicroRNA profiling in prostate cancer—the diagnostic potential of urinary miR-205 and miR-214. PLoS ONE. 2013;8:e76994. doi: 10.1371/journal.pone.0076994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rooij E, Kauppinen S. Development of micro RNA therapeutics is coming of age. EMBO Mol Med. 2014;6:851–864. doi: 10.15252/emmm.201100899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kolijn K, Verhoef EI, Smid M, Bottcher R, Jenster GW, Debets R, et al. Epithelial–mesenchymal transition in human prostate cancer demonstrates enhanced immune evasion marked by IDO1 expression. Cancer Res. 2018;78:4671–4679. doi: 10.1158/0008-5472.CAN-17-3752. [DOI] [PubMed] [Google Scholar]
- 172.Brase JC, Johannes M, Schlomm T, Haese A, Steuber T, Beissbarth T, et al. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer. 2011;128:608–616. doi: 10.1002/ijc.25376. [DOI] [PubMed] [Google Scholar]
- 173.Cheng HH, Mitchell PS, Kroh EM, Dowell AE, Chéry L, Siddiqui J, et al. Circulating microRNA profiling identifies a subset of metastatic prostate cancer patients with evidence of cancer-associated hypoxia. PLoS ONE. 2013;8:e69239. doi: 10.1371/journal.pone.0069239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kelly B, Miller N, Sweeney K, Durkan G, Rogers E, Walsh K, et al. A Circulating MicroRNA signature as a biomarker for prostate cancer in a high risk group. J Clin Med. 2015;4:1369–1379. doi: 10.3390/jcm4071369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Quirico L, Orso F. The power of microRNAs as diagnostic and prognostic biomarkers in liquid biopsies. Cancer Drug Resist. 2020;3:117–139. doi: 10.20517/cdr.2019.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hao Y, Zhao Y, Zhao X, He C, Pang X, Wu TC, et al. Improvement of prostate cancer detection by integrating the PSA test with miRNA expression profiling. Cancer Invest. 2011;29:318–324. doi: 10.3109/07357907.2011.554477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Luu HN, Lin HY, Sørensen KD, Ogunwobi OO, Kumar N, Chornokur G, et al. miRNAs associated with prostate cancer risk and progression. BMC Urol. 2017;17:18. doi: 10.1186/s12894-017-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Petrylak DP, Tangen CM, Hussain MHA, Lara PN, Jones JA, Taplin ME, et al. Docetaxel and estramustine compared with mitoxantrone and prednisone for advanced refractory prostate cancer. N Engl J Med. 2004;351:1513–1520. doi: 10.1056/NEJMoa041318. [DOI] [PubMed] [Google Scholar]
- 179.Tannock IF, De Wit R, Berry WR, Horti J, Pluzanska A, Chi KN, et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med. 2004;351(15):1502–1512. doi: 10.1056/NEJMoa040720. [DOI] [PubMed] [Google Scholar]
- 180.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 181.Altieri DC, Languino LR, Lian JB, Stein JL, Leav I, Van Wijnen AJ, et al. Prostate cancer regulatory networks. J Cell Biochem. 2009;107:845–852. doi: 10.1002/jcb.22162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.