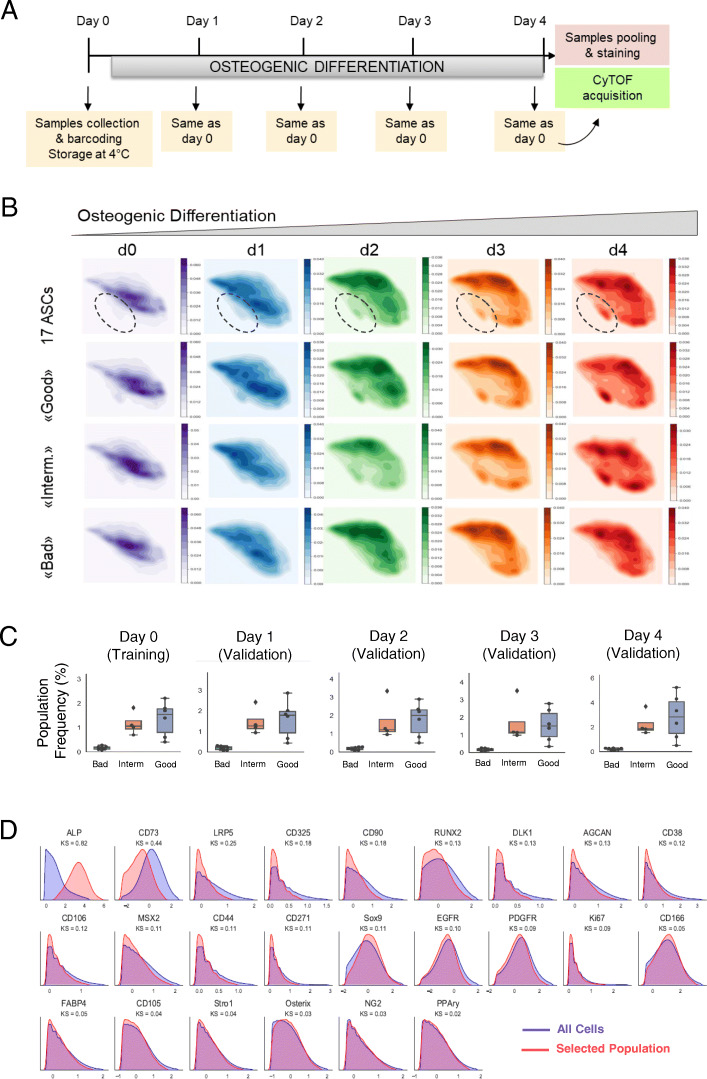
Fig. 3.
Identification of AD-MSC osteogenic subpopulation. a Sample collection and CyTOF approach scheme during 4 days of osteogenic differentiation (d0 = undifferentiated state, d1–d4 = differentiation). b Cell density plots on the UMAPs of the five analyzed days (d0, d1, d2, d3, d4) during osteogenic differentiation. Once the pool of all 17 AD-MSC lines is represented, once only the “good,” the “intermediate” (interm.), and the “bad” AD-MSC lines. Highlighted is the emerging population during osteogenic differentiation. Bright color indicates lower density, and dark color indicates higher cellular density. c Empirical distribution densities of all analyzed 31 marker abundances for the entire cell population (blue) and the cell subset selected by CellCNN (red). The identified subpopulation is characterized by alkaline phosphatase-positive (ALP+) and CD73low expressing cells. d Boxplots indicating the frequencies of the ALP+/CD73low subpopulation selected by CellCNN in all “good,” “intermediate” (interm.), and “bad” osteogenic differentiating lines during the five analyzed days. Error bars represent the mean of the percentage of positive cells present in “good” (n = 6), “intermediate” (n = 4), and “bad” (n = 7) AD-MSCs