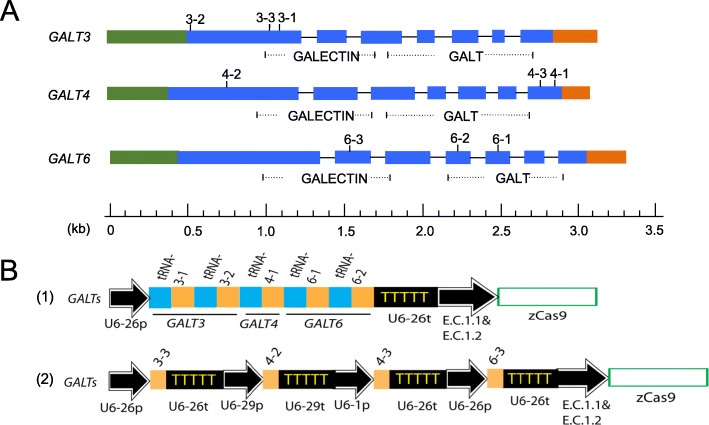
Fig. 1.
Schematic illustration of gRNA target sites and the two CRISPR multiplexing constructs. a. Three gRNAs were chosen for each of the GALTs, which belong to the five-membered galactosyltransferase (GALT) gene family. gRNAs were labeled as 3–1, 3–2, and 3–3 for GALT3; 4–1, 4–2, and 4–3 for GALT4; 6–1, 6–2, and 6–3 for GALT6; these gRNAs were used in gene constructs shown in 1b. (Green and orange represent 5′-UTR and 3′-UTR regions, respectively; blue represents exons; black lines represent introns. Online software named CRISPR-P 2.0 (http://crispr.hzau.cn/cgi-bin/CRISPR2/CRISPR) was used to design all gRNAs. Pfam domain predictions: Pf01762 corresponds to the Galactosyltransferase (GALT) domain; Pf00337 corresponds to the Galactose-binding lectin (GALECTIN) domain. b-1. Five gRNAs (3–1, 3–2, 4–1, 6–1, and 6–2 shown in 1a) with each gRNA fused with a tRNA were cloned in a single polycistronic transcription unit to target GALT3, GALT4, and GALT6 simultaneously. b-2. Four gRNAs (3–3, 4–2, 4–3, and 6–3 in 1a) were assembled as four individual transcription units to target GALT3, GALT4, and GALT6. Both B-1 and B-2 were cloned in the pHEE401E vector, which contains a maize codon-optimized Cas9 gene (zCas9) driven by an Arabidopsis egg-cell specific promoter (E.C.1.1) fused with an egg-cell specific enhancer (E.C.1.2) [19]