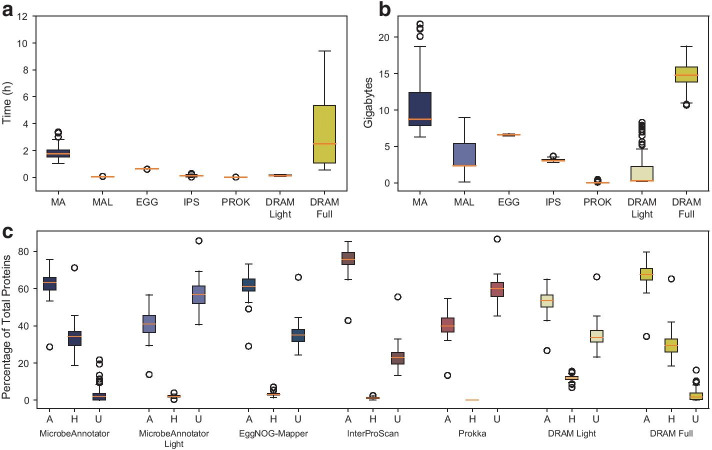
Fig. 3.
Annotation tool comparison using CPR bacteria genomes. Given the smaller genome sizes and lower completeness levels, all tools required shorter times to perform annotations (a). While the standard mode of MicrobeAnnotator only takes ~ 2 h to annotate a genome, the light version takes only a couple of minutes like other tools that use smaller databases. Consequently, the RAM usage for these smaller genomes decreases but is still on par with DRAM requirements (b). As expected for genomes with lower representation in the databases, all tools could annotate a lower percentage of proteins. However, MicrobeAnnotator was able to find more matches for hypothetical proteins, having one of the lowest percentages of unannotated proteins of all tools (c). MA MicrobeAnnotator, MAL MicrobeAnnotator Light, EGG EggNOG-Mapper, IPS InterProScan, PROK Prokka. A: Annotated, H: Hypothetical, U: Unannotated