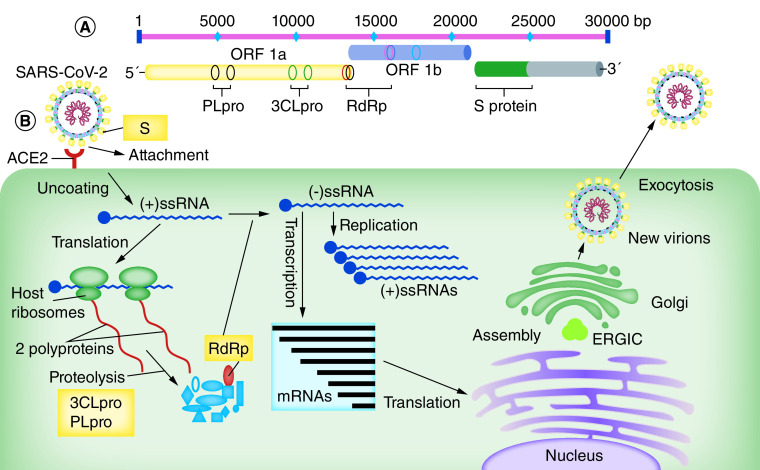
Figure 1. The genome organization and replication cycle of the SARS-CoV-2.
(A) Genome analysis of SARS-CoV-2 reveals that the genome length is approximately 30 kb with several ORFs. Viral structural proteins such as 3CLpro, PLpro and RdRp are encoded from the two main ORFs. (B) Coronavirus infection begins with the attachment of the spike protein (S) with its receptor on the host cell’s surface (ACE2). Following adsorption and uncoating of the virus inside the host cells with an endosomal pathway, and releasing the NC to the cytoplasm, the viral ssRNA genome attaches to the host ribosomes to produce two polyproteins that are subsequently cleaved (in a proteolysis pathway) into smaller components, by host and viral proteases including 3CLpro and PLpro. The RdRp is involved in virus genome replication and also in viral mRNAs and their relevant viral proteins production. Then assembling of viral genome and viral proteins into new virions occurs in the host cell ERGIC. Eventually, new virions are transmitted outside the cells through exocytosis.
3CLpro: 3-Chymotrypsin-like protease; ACE2: Angiotensin-converting enzyme 2; ERGIC: Endoplasmic reticulum-Golgi intermediate compartment; NC: Nucleocapsid; ORF: Open-reading frame; PLpro: Papain-like protease; RdRp: RNA-dependent RNA polymerase; S: Spike protein; SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2.