Abstract
Abdominal aortic aneurysm (AAA), when ruptured, results in high mortality. The identification of molecular pathways involved in AAA progression is required to improve AAA prognosis. The aim of the present study was to assess the key genes for the progression of AAA and their functional role. Genomic and clinical data of three independent cohorts were downloaded from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) (GSE57691, GSE7084, and GSE98278). To develop AAA diagnosis and progression-related differentially expressed genes (DEGs), we used a significance analysis of microarray (SAM). Spearman correlation test and gene set analysis were performed to identify potential enriched pathways for DEGs. Only the Frizzled-related protein (FRZB) gene and chromosome 1 open reading frame 24 (C1orf24) exhibited significant down-regulation in all analyses. With FRZB, the pathways were associated with RHO GTPase and elastin fiber formation. With C1orf24, the pathways were elastic fiber formation, extracellular matrix organization, and cell–cell communication. Since only FRZB was evolutionally conserved in the vertebrates, function of FRZB was validated using zebrafish embryos. Knockdown of frzb remarkably reduced vascular integrity in zebrafish embryos. We believe that FRZB is a key gene involved in AAA initiation and progression affecting vascular integrity.
Keywords: abdominal aortic aneurysm, bioinformatics, FRZB, gene expression omnibus, vascular integrity, zebrafish
Introduction
Abdominal aortic aneurysm (AAA) is a late age-of-onset disorder that affects approximately 5% of men aged 65–74 years; when ruptured, AAAs are associated with a mortality rate of up to 90% [1]. Ruptured aortic aneurysms are a major cause of death, and they were reported to be the 13th leading cause of mortality in 2017 [2]. Preemptive elective open surgical or endovascular repair is considered when the AAA diameter is ≥55 mm, when it is rapidly expanding (≥10 mm/year), or when it causes symptoms [3]. There are, however, large intrapatient and interpatient variations with regard to rates of expansion of small AAAs during follow-up [4]. Therefore, increasing demand to improve risk stratification in patients with AAA has been claimed [5,6].
The pathophysiology of AAA has been extensively studied. AAA was previously believed to be a form of atherosclerosis; however, it is now recognized as a distinct degenerative process involving all layers of the vessel wall and is characterized by destruction of elastin and collagen in the media and adventitia and loss of smooth muscle cells with thinning of the media [1,7,8]. However, a lack of knowledge remains regarding the molecular mechanisms that initiate aneurysm formation and expansion. In current clinical practice, the strategy to stratify patients at high risk for aneurysm progression is lacking.
Serial monitoring of biological activity of AAA would be necessary to lower mortality and morbidity from AAA rupture; however, no biomarkers have yet proven to be of prognostic value additive to AAA diameter to predict AAA expansion [9,10]. AAA growth rate can only be acknowledged retrospectively. To be able to find preventive strategies, the molecular mechanisms behind the disease and drug targets need to be evaluated in detail.
Some studies have attempted to evaluate the candidate genes and pathways potentially associated with AAA, but a large portion of their data was prone to bias and focused on pre-selected genes encoding matrix metalloproteinases, components of the immune system, and known atherosclerosis-related factors [8,11–14]. Therefore, to understand the molecular pathways controlling the progression of AAA without bias, we examined the statistical analysis of three cohorts acquired from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) Series (GSE57691, GSE7084, and GSE98278).
Methods
Patient data
Microarray gene expression data and clinical information of three different AAA patient cohorts were obtained from three independent sources: James Cook University, Wayne State University School of Medicine, and Ludwig Maximilians University Munich. They were accessed through the NCBI GEO Series (GSE57691, GSE7084, and GSE98278, respectively). GSE57691 and GSE7084 contain both normal and AAA specimen data. Thus, they were evaluated to find the differentially expressed genes (DEGs) among the AAA patient group. On the contrary, GSE98278, which does not include the normal patient data, was used to determine genes that are linked to the further development of AAA. More details of the patient data are shown below (Table 1). Since the data we used were publicly accessible, IRB approval was not required.
Table 1. Patients’ characteristics.
| GSE57691 | GSE7084 | GSE98278 | ||||||
|---|---|---|---|---|---|---|---|---|
| Non-AAA | AAA | Non-AAA | AAA | Size | Status | |||
| Intermediate | Large | Stable | Rupture | |||||
| n | 10 | 49 | 11 | 10 | 15 | 16 | 31 | 17 |
| Age (years) | 68.4 (±4.5) | 69.8 (±7.0)* | 64.5 (±10.7) | 66.4 (±5.7) | 67.7(±7.4) | 72.3(±9.2) | 69.5 (±7.2) | 73.5 (±11.3) |
| Women (%) | 40.0% | 4.1% | 27.3% | 20.0% | 6.7% | 12.5% | 3.2% | 23.5% |
| Diameter (mm) | - | 62.3 (±10.2)* | - | - | 54.1 (±1.8) | 84.1 (±12.6) | 62.3 (±12.1) | 77.0 (±14.7) |
| Hypertension (%) | - | 81.6% | - | - | 86.7% | 93.8% | 93.5% | 88.2% |
| Diabetes (%) | - | 22.4% | - | - | 26.7% | 31.3% | 32.3% | 23.5% |
| Dyslipidemia (%) | - | 71.4% | - | - | 86.7% | 81.3% | 77.4% | 82.3% |
| Coronary heart disease (%) | - | 51.0% | - | - | 46.7% | 43.8% | 58.1% | 41.2% |
| Ever smoker (%) | 50.0% | 57.1% | - | - | 33.3% | 68.8% | 58.1% | 58.8% |
Nominal variables are presented as percentages; continuous variables are presented as mean ± standard deviation. Asterisk (*) means Cohen’s formula for pooled standard deviation.
Standardization
In microarray data, each gene has a different range of expression values. For instance, while some gene expression levels range from 0 to 100, others can range from 500 to 1000. Thus, to facilitate comparison between them, we had to standardize their values and to bring all the genes to the same range. Since microarray data from GSE57691 had been distributed after standardization, only gene expression measurements from GSE7084 and GSE98278 needed to be standardized. The standardization process has assigned a standard score (z-score) to each gene expression value in the data. The ‘scale’ function in the ‘base’ package R (version 3.6.1) was used in this step.
Missing values
There were no missing values from GSE57691, GSE7084, and GSE98278. However, a huge portion of genes within the three datasets appeared repetitively in different columns. In order to organize these recurring genes, we aggregated the genes in the columns by mean, using the ‘aggregate’ function in the ‘stats’ package R (version 3.6.1). In consequence, a large number of columns (14190, 3023, and 15893) from each gene expression dataset were dropped, but the total number of distinct gene symbols has not changed and genetic significance also remains the same.
Significance analysis of microarray
Significance analysis of microarray (SAM) is a test method where the test statistic measures the difference in gene-expression values between control and treatment groups divided by the standard deviation of repeated measurements. It is specifically designed to address data from large microarrays, as SAM guarantees the probability of error to not increase with the number of comparisons, in contrast with an increase observed with the number of t tests. Once all test statistic scores are acquired, they are ranked, and genes with scores greater than an adjustable threshold delta (Δ) are selected. Subsequently, the false discovery rate (FDR) is computed to identify the rate of genes incorrectly identified as significant by using the permutations of the measurements [16]. In this study, we performed the SAM tests in the ‘siggenes’ package R (version 3.6.1). Total permutations (1000) and FDR = 0.0005 were used for both cohorts.
Additional t test and correlation analysis
Using the GSE98278 dataset which contains stability (stable vs. ruptured) and size (intermediate vs. large) information of AAA, we next aimed to find out the DEGs that are actively engaged in the development of AAA. Unpaired t tests were implemented to 12 commonly significant genes from two previous SAM tests, and the resulting P-values were corrected with Benjamini–Hochberg procedure (FDR = 0.25). Additionally, we conducted correlation tests between two genes of our main focus, Frizzled-related protein (FRZB) and chromosome 1 open reading frame 24 (C1orf24), and the other genes in each cohort (GSE7084, GSE57691) for gene set analysis. |R| > 0.7 and P<0.05 were used as cut-off values. To obtain the signaling pathways, correlated genes with FRZB and C1orf24 were subjected to Reactome pathway analysis using Enrichr (https://amp.pharm.mssm.edu/Enrichr/) [15].
Maintenance of zebrafish and morpholino injection
Wild-type AB zebrafish and fli1; DsRed transgenic zebrafish were maintained in an automatic circulation system (Genomic-Design, Daejeon, Korea) at 28.5°C [16]. Every experiment using zebrafish embryos were performed in accordance with the guidelines of the Ulsan National Institute of Science and Technology (UNIST) Institutional Animal Care and Use Committee (IACUC) (IACUC approval number: UNISTIACUC-15-14, date: 2016-10-11) at UNIST building 103. Zebrafish embryos for experiments were cultured using E3 solution in incubators at 28°C. A translation-blocking morpholino (MO) targeting frzb (Gene Tools, Philomath, U.S.A.) was resolved in DEPC water in 25 ng/nl stock. The sequence of frzb ATG MO is 5′-AGCGGAGTTGATAGAAGAATGACAT-3′. The morpholino targeting frzb was injected in embryos of wild-type AB zebrafish at the one-celled stage of development. Microinjections were performed using Femtojet 4i microinjector (Eppendorf, Hamburg, Germany).
Confocal imaging for the live zebrafish embryo
The zebrafish embryos were anesthetized using 0.02% tricaine and mounted with 3% methyl cellulose for imaging. Then, embryos were observed with a confocal microscope (LSM880, Carl Zeiss, Oberkochen, Germany).
Results
Data characteristics
To evaluate candidate genes of progression of AAA, we analyzed three cohorts, specifically GSE57691, GSE7084, and GSE98278. For GSE57691, gene expression was assessed in aortic biopsies from ten patients that were organ donors non-AAAs and 49 patients diagnosed with AAAs (mean AAA diameter: 62.3 ± 10.2 mm) (Table 1). AAA patients were less likely to be female compared with the non-AAA patients (small AAA: 0%, large AAA: 7%, non AAA: 40%; P<0.05). For GSE7084, aortic wall samples were collected from 10 AAA patients who underwent repair operation for AAA and from 11 non-AAA autopsies. The non-AAA samples from GSE57691 and GSE7084 were defined an abdominal aorta with size of under 50 mm. Of the ten patients experiencing AAA, four samples were used for further analysis along with another four aortic samples acquired from the non-AAA cohort. These were all clustered as a pool in the raw data using average statistics. Specifically, we disassembled the two pools to allow for a correct representation of the sample size. For GSE98278, the cohort consisted of 48 patients undergoing elective (stable AAA, n=31) and emergency (ruptured AAA, n=17) treatment. The stable AAA and the ruptured AAA groups exhibited differences with regard to age (69.5 ± 7.2 vs 73.5 ± 11.3; P<0.05), sex (3 vs 23% female sex; P<0.05), and AAA diameter (62.3 ± 12.1 vs 77.0 ± 14.7; P<0.05). Within the stable AAA group, the patients were divided according to the size of their AAAs into intermediate (54.1 ± 1.8 mm) and large (84.1 ± 12.6 mm) subgroups.
Identification of genes associated with AAA progression
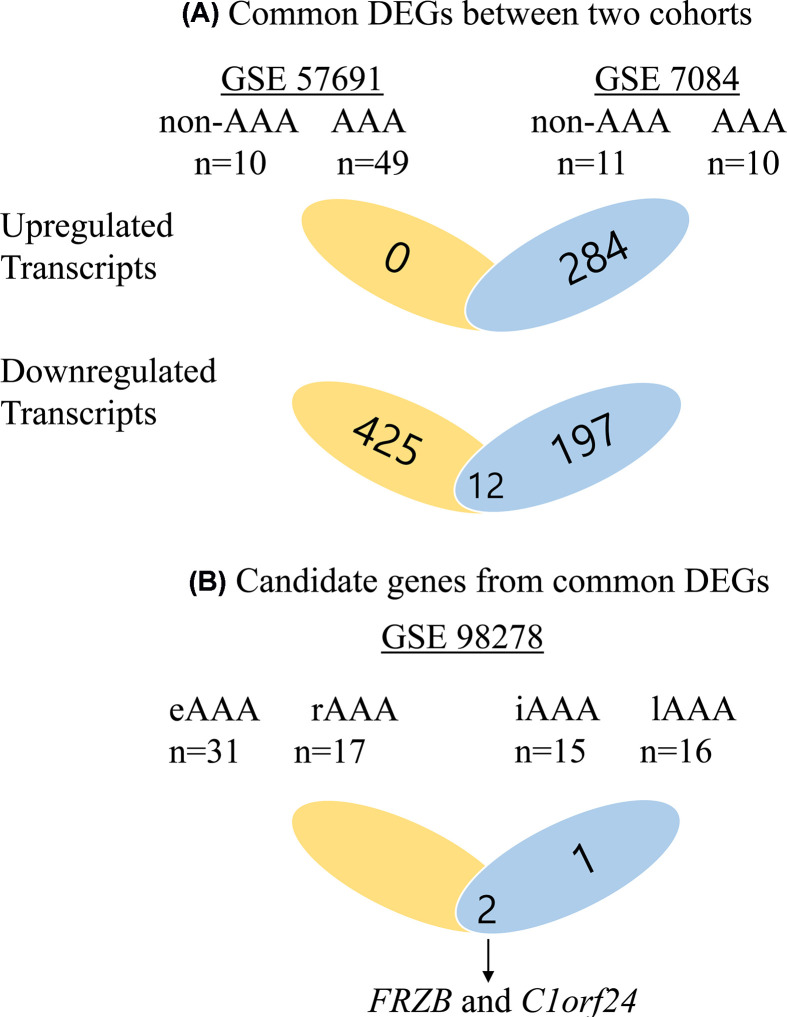
We performed SAM to identify DEGs within each cohort, and the study cohorts included GSE57691, GSE7084 divided into two groups, non-AAAs and AAAs. A total of 436 down-regulated genes were identified from GSE57691, and 284 up-regulated and 209 down-regulated genes were identified from GSE7084 (Figure 1). A total of 12 DEGs were statistically significant in both cohorts. DEGs are listed in Supplementary Table S1. Of these 12 genes, t test analysis was performed on GSE98278 to show the difference of expression in two subgroups, intermediate size and large size and stable AAA and ruptured AAA. Three genes (C1orf24, FRZB, and KIAA0367) and two genes (C1orf24 and FRZB) were differentially expressed at the first subgroup and second subgroup, respectively. C1orf24 and FRZB were shown to be simultaneously altered in a statistically significant manner as the size and instability of AAA increased (Figure 1). The mRNA expression of the C1orf24 and FRZB genes was decreased in AAA patients from GSE57691 and 7081 (Figure 2A,B) and with increasing size and instability of AAA from GSE982789 (Figure 2C,D).
Figure 1. Flow charts of study design.
(A) Up-regulated transcripts were 0 from GSE57691 and 284 from GSE7084. Down-regulated transcripts were 438 from 57691 and 208 from GSE7084. Twelve genes were significant in both cohorts. (B) Of these 12 genes, 3 were significant between intermediate size of AAA and large size of AAA in GSE 98278; 2 were significant between stable AAA and rupture AAA in GSE 98278. FRZB and C1orf24 were concomitantly significant. Abbreviations: eAAA, elective AAA; iAAA, intermediate AAA; lAAA, large AAA; rAAA, ruptured AAA.
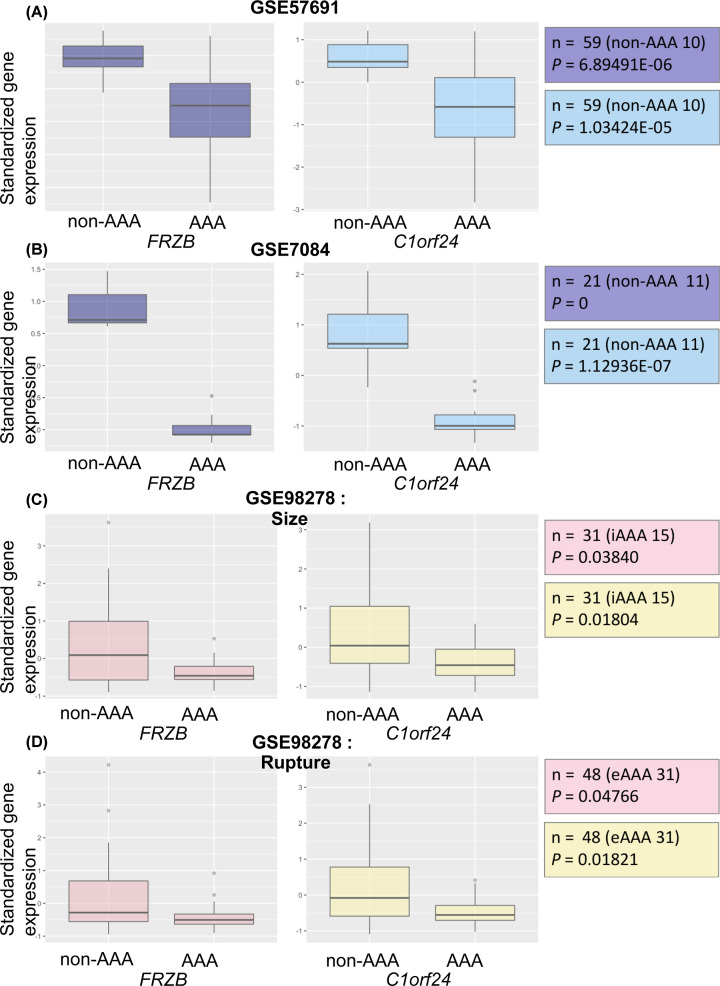
Figure 2. Box plots for the expression values of FRZB and C1orf24.
(A) The expression values of between FRZB and C1orf24 between AAA patients (n=49) and non-AAA patients (n=10) from GSE 57691. FRZB and C1orf24 were significantly down-expressed in AAA patients. (B) The expression values of FRZB and C1orf24 between AAA patients (n=10) and non-AAA patients (n=11) from GSE 7084. FRZB and C1orf24 were significantly down-expressed in AAA patients. (C) The expression values of FRZB and C1orf24 between intermediate size AAA (n=15) and large size AAA (n=16) from GSE 98278. FRZB and C1orf24 were significantly down-expressed in large size AAA. (D) The expression values of FRZB and C1orf24 between eAAA (n=31) and rAAA (n=17) from GSE 98278. FRZB and C1orf24 were significantly down-expressed in ruptured AAA. Abbreviations: eAAA, elective AAA; iAAA, intermediate AAA; lAAA, large AAA; rAAA, ruptured AAA.
Correlation analysis
To identify the pathway correlated with FRZB and C1orf24, we performed Pearson’s correlation of FRZB and C1orf24 with other genes GSE57691 and GSE7084. By applying |R| > 0.7 and P<0.05, 64 genes were positively correlated with FRZB simultaneously from GSE57691 and GSE7084. Seventy-one genes were positively correlated with C1orf24, and three genes were negatively correlated with C1orf24 from two cohorts (Supplementary Table S2). Correlated genes with FRZB and C1orf24 were subjected to Reactome pathway analysis using Enrichr. With FRZB, the pathways were associated with RHO GTPase and elastin fiber formation (Table 2). With C1orf24, the pathways were elastic fiber formation, extracellular matrix organization, and cell–cell communication (Table 3).
Table 2. Top ten significantly enriched Reactome pathways with FRZB and correlated genes from GSE57691 and GSE7084 using Enrichr (https://amp.pharm.mssm.edu/Enrichr/).
| Term | Overlap | P-value | Odds ratio | Combined score | Genes |
|---|---|---|---|---|---|
| RHO GTPases activate ROCKs | 3/17 | 2.16E-05 | 54.29864253 | 583.4084401 | PPP1CB; MYH11; MYH10 |
| RHO GTPases activate PAKs | 3/21 | 4.18E-05 | 43.95604396 | 443.2037946 | PPP1CB; MYH11; MYH10 |
| Molecules associated with elastic fibers | 3/30 | 1.25E-04 | 30.76923077 | 276.5468079 | MFAP4; ITGA8; FBLN5 |
| Elastic fiber formation | 3/41 | 3.20E-04 | 22.51407129 | 181.1923658 | MFAP4; ITGA8; FBLN5 |
| RHO GTPases activate PKNs | 3/60 | 9.82E-04 | 15.38461538 | 106.5481366 | PPP1CB; MYH11; MYH10 |
| RHO GTPases activate CIT | 2/16 | 0.001211888 | 38.46153846 | 258.2913719 | MYH11; MYH10 |
| Semaphorin interactions | 3/67 | 0.001353045 | 13.77726751 | 91.00432898 | ITGA1; MYH11; MYH10 |
| Integrin cell surface interactions | 3/67 | 0.001353045 | 13.77726751 | 91.00432898 | ITGA1; ITGA8; JAM3 |
| Extracellular matrix organization | 5/283 | 0.002262817 | 5.436259853 | 33.11304623 | MFAP4; ITGA1; ITGA8;FBLN5; JAM3 |
| Sema4D induced cell migration and growth-cone collapse | 2/24 | 0.00274105 | 25.64102564 | 151.2670332 | MYH11; MYH10 |
Table 3. Top ten significantly enriched Reactome pathways with C1orf24 and correlated genes from GSE57691 and GSE7084 using Enrichr (https://amp.pharm.mssm.edu/Enrichr/).
| Term | Overlap | P-value | Odds ratio | Combined score | Genes |
|---|---|---|---|---|---|
| Elastic fiber formation | 3/41 | 4.32E-04 | 20.32520325 | 157.4485645 | MFAP4; EFEMP1; LOXL4 |
| Extracellular matrix organization | 6/283 | 5.44E-04 | 5.889281508 | 44.26820723 | MFAP4; EFEMP1; ITGA1; LOXL4; COL8A1; JAM3 |
| Cell–cell communication | 4/131 | 0.001281745 | 8.481764207 | 56.48458548 | CLDN12; CTNNA1; CDH13; WASL |
| Cell–cell junction organization | 3/61 | 0.001386 | 13.6612 | 89.90722 | CLDN12; CTNNA1; CDH13 |
| RHO GTPases activate ROCKs | 2/17 | 0.001678 | 32.67974 | 208.8211 | PPP1R12A; MYH10 |
| Semaphorin interactions | 3/67 | 0.001817 | 12.43781 | 78.49063 | DPYSL3; ITGA1; MYH10 |
| RHO GTPases activate PAKs | 2/21 | 0.002568 | 26.45503 | 157.7983 | PPP1R12A; MYH10 |
| Cell junction organization | 3/86 | 0.003697 | 9.689922 | 54.26647 | CLDN12; CTNNA1; CDH13 |
| Molecules associated with elastic fibers | 2/30 | 0.005209 | 18.51852 | 97.35994 | MFAP4; EFEMP1 |
| Adherens junctions interactions | 2/31 | 0.005555 | 17.92115 | 93.06567 | CTNNA1; CDH13 |
frzb is essential for the development of zebrafish embryo through regulation of vascular integrity
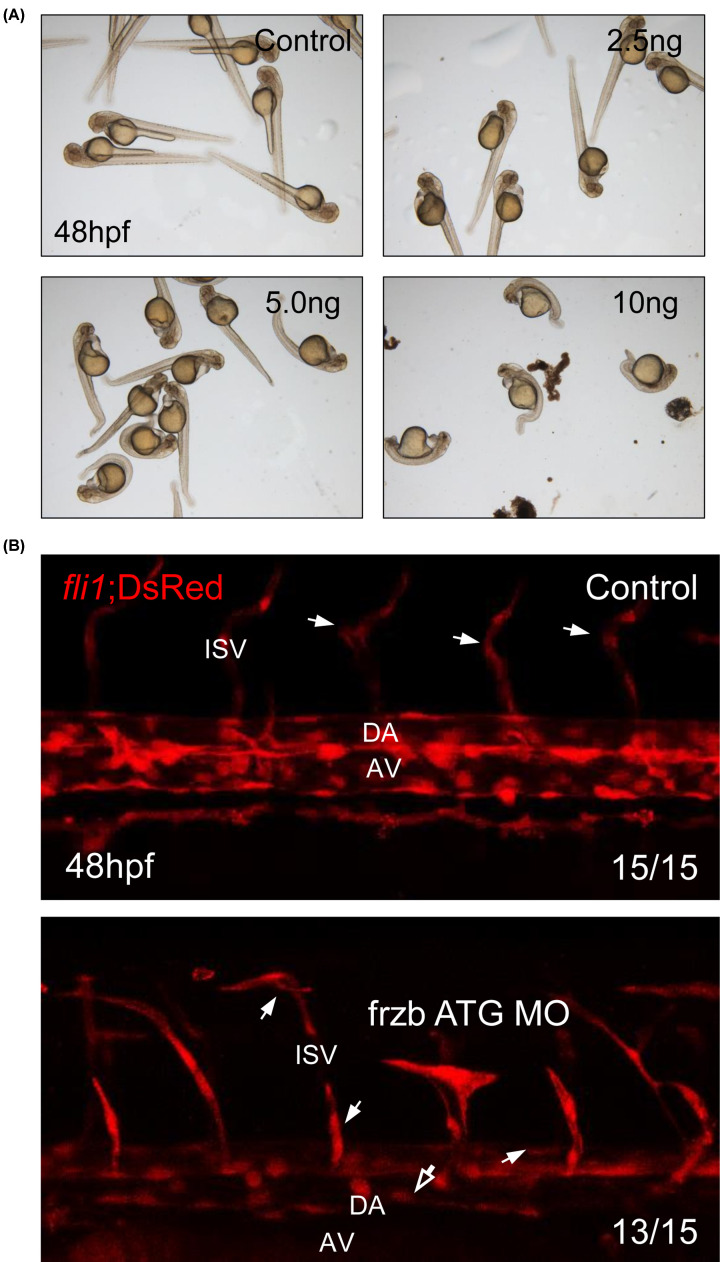
RHO GTPase pathway and elastic fiber formation-related genes are analyzed from correlation analysis, and those genes are associated with vascular integrity. C1orf24 was not evolutionally conserved in the vertebrates, whereas FRZB was conserved. Alignment between human FRZB and zebrafish frzb shows 70% exactly same residue, 12% similar structure of residue, and 6% gaps in compositional matrix adjustment (Supplementary Figure S1). Considering these data, we validated our correlation data from FRZB by observing vascular formation of zebrafish embryos. To investigate the function of frzb in vascular integrity, knockdown of frzb was processed using MO injection. Dependent on dosage of frzb ATG MO, development of zebrafish embryos was interrupted (Figure 3A). Since development of embryos with 5 and 10 ng were highly interrupted and developmental defects were shown from 2.5 ng of MO-njected group, we decided 2.5 ng is enough for knockdown.
Figure 3. frzb is essential for the development of zebrafish embryo through regulation of vascular integrity.
(A) Imaging of live embryos after morpholino injection (0, 2.5, 5, and 10 ng). (B) Confocal imaging of fli1; DsRed transgenic zebrafish embryos from control embryos and frzb ATG MO-injected embryos. Abbreviations: AV, axial vein; DA, dorsal aorta; hollow arrow, narrow dorsal aorta and axial vein; ISV, intersomite vessel. White arrow, abnormal branch of intersomite vessel.
Frzb is highly correlated with elastic fiber formation-related genes at patient database (Table 2), and elastic fiber is important for the vascular integrity. To validate our informatics data, vascular formation of zebrafish embryos was checked using fli1;DsRed transgenic zebrafish. As many zebrafish embryos with defects in vascular integrity showed abnormal vascular formation [17,18], frzb ATG MO-injected embryos show disrupted vascular structure in trunk. Lumen of dorsal aorta and axial vein became narrow, and abnormal branch of intersomite vessels were observed (Figure 3B). These data suggest that frzb is essential for the vascular integrity.
Discussion
Even though multiple etiological, genetic factors contributing to AAA, as well as its pathobiology, have been extensively studied, the mechanisms of progression of AAA are still incompletely understood. Through analysis of three gene expression studies, we tried to narrow down possible candidate genes that provide a comprehensive view of the disease.
Through literature review, we had found three articles which analyzed DEGs of AAA from datasets in GEO [19–21]. The datasets used from each articles were as follows; Shen et al. [19], GSE47472; Wan et al. [20], GSE7084, GSE47472, and GSE57691; Chen et al. [21], GSE7084, GSE47472, and GSE57691. The tissue samples in GSE47472 were non-aneurysmal aortic neck tissues, so we considered the data in GSE47472 were not suitable for our study design. Because the data in GSE98278 included important clinical data such as size of AAA and rupture status of AAA, we analyzed the data of GSE98278 with other two datasets, GSE7084 and GSE57691.
By applying very low FDR value (0.0005) in analysis of DEGs from two cohorts (GSE7084 and GSE57691), only 12 genes were identified. The major limitation of microarray-based mRNA expression profiling is that tissue samples from end-stage disease provide little information about initiation [22]. The only option for vascular surgeons to retrieve tissue samples of AAA is through open surgeries of AAA, in which diameter is mostly over 50 mm. In our study, the mean diameter of AAA from GSE57691 is 62.3 mm. By comparing disease-progressed tissue samples and normal tissues, it is non-logical to develop DEGs involving initiation of AAA. Therefore, 12 DEGs from GSE 7084 and GSE 57691 were analyzed to identify which genes were involved in terminal AAA and rupture. Only two genes, FRZB and C1orf24, were differentially expressed along size expansion and rupture. FRZB binds to both Wnt-8 and Wnt-1 and acts as a functional inhibitor of Wnt-8 activity in Xenopus embryos [23]. FRZB has been reported as a key factor of skeletal morphogenesis and osteoarthritis [24,25]. C1orf24 contains a DnaJ motif, a feature of heat shock protein that was investigated as a factor associated with renal carcinogenesis [26]. It was also reported that C1orf24 was up-regulated in thyroid cancer and might increase proliferation and cell migration [27].
To evaluate which of the molecular pathways were related to FRZB and C1orf24, we performed correlation analysis on GSE 7084 and GSE57691. Then, pathway analysis with correlated genes revealed correlated genes with FRZB and C1orf24 genes are involved in the pathway of RHO GTPase and elastin fiber formation. In many experimental studies, RHO GTPase played central roles in vascular smooth muscle cell proliferation, migration, and contractility, differentiation, and ROS generation [28–32]. In rats with balloon‐injured arteries, increased RhoA/ROCK activity contributes to neointimal formation, and these detrimental effects are significantly suppressed by the ROCK‐specific antagonist Y27632 [33]. Regarding VSMC differentiation, through modulating serum response factor-dependent skeletal and smooth muscle gene expression, the RhoA/ROCK signaling pathway regulates VSMC differentiation [34]. The important characteristics of aneurysmal wall are breakdown of elastin and collagen, VSMC apoptosis, and activation of immune responses [35]. We showed FRZB and C1orf24 correlated with RHO GTPase and elastin fiber formation pathways. Among them, we validated that FRZB is essential for the vascular integrity using zebrafish embryos. That means low expression of FRZB is associated with the progression of AAA by diminishing integrity of abdominal aorta.
Many studies have validated the candidate genes of AAA pathobiology through hypothesis-driven way. These studies have been on a ‘best guess’ basis and do not fit with the studies of diseases for which underlying pathological processes are unclear [36]. With advances in genetic technology and bioinformatics, unbiased high throughput genome-wide association studies (GWASs) have widely made. In meta-analysis of GWASs for AAA, four new AAA risk loci, rs1795061 (SMYD2), rs9316871 (LINC00540), rs3827066 (PCIF1/MMP9/ZNF335), and rs2836411 (ERG), were identified, as well as five of the six previous AAA genetic associations, rs602633 (PSRC1-CELSR2-SORT1), rs4129267 (IL6R), rs10757274 (CDKN2BAS1/ANRIL), rs10985349 (DAB2IP), and rs6511720 (LDLR) [37]. One study used VSMCs isolated from patients with AAA and controls to assess DNA methylation in the genes found within the nine associated loci, and altered DNA methylation levels were found in three genes (ERG, IL6R, and SMYD2) [38]. Despite their highly significant associations with AAA, these genetic loci explain only a small proportion of the heritability of AAA.
Even though microarray-based mRNA expression profiling has limitations described before and difficulties, it can a useful tool in dissecting disease pathogenesis, especially when pathogenesis of disease is not well established. Among three cohorts we analyzed, Lenk et al. (GSE 7084) compared whole genome expression profile between AAA (n=10) and normal (n=10) and showed broad coordinate gene expression in immunological pathways [39]. FRZB was one of the top 100 most differential genes in Supplementary Table S2. Biros et al. (GSE 57691) used AAA tissue samples for analyzing expression difference between AAA and aortic occlusive disease [40]. Direct comparison of our study with GSE57691 is impossible; however, FRZB and C1orf24 was down-regulated in AAA group. Gäbel et al. (GSE98278) tried to validate the candidate genes relating to terminal AAA [41]. Based on the studies that molecular mechanisms and genetic factors might differ in initiation, growth, and rupture of AAA [42], they developed ten DEGs and showed that these genes converged at activation of HIF-1α network. They also showed histologic quantification of angiogenesis in ruptured AAA. Even though it might provide important insight of terminal AAA, by not comparing normal aortic tissue, genes involving process of AAA development might be neglected. Therefore, we tried to narrow down the genes comparing AAA tissues and normal aortic tissue, then analyzed on two groups, size difference and rupture. We considered this analytic strategy is more logical to develop candidate genes of AAA progression.
To prove the protein level of candidate genes in AAA patients, the optimal tissue for our purpose was aneurysmal tissue retrieved from open surgery of AAA. In the Biobank of our country, however, there was no tissue sample of AAA. This is our major limitation. Further investigations will be held to validate the correlation of these pathways with our candidate genes.
Conclusions
The present study examined the candidate genes involved in the progression of AAA. We were able to identify two gene expressions (FRZB and C1orf24) by analyzing three cohorts. In pathway analysis with correlated genes with these two candidate genes (FRZB and C1orf24), we showed the pathways were related to VSMC and elastin fiber formation, which might imply these candidate genes are involved in aneurysm integrity. Additionally, we validated that FRZB was linked to the vascular integrity using zebrafish embryos. Considering our findings, we believe that FRZB is a possible candidate gene involved in AAA progression.
Supplementary Material
Abbreviations
- AAA
abdominal aortic aneurysm
- C1orf24
chromosome 1 open reading frame 24
- DEG
differentially expressed gene
- DEPC
diethylpyrocarbonate
- FDR
false discovery rate
- FRZB
Frizzled-related protein
- GEO
Gene Expression Omnibus
- GWAS
genome-wide association study
- HIF-1α
hypoxia-inducible factor 1 alpha
- IACUC
Institutional Animal Care and Use Committee
- MO
morpholino
- NCBI
National Center for Biotechnology Information
- ROCK
Rho-associated protein kinase
- SAM
significance analysis of microarray
- UNIST
Ulsan National Institute of Science and Technology
- VSMC
vascular smooth muscle cell
Contributor Information
Jin Mo Kang, Email: calzevi@gmail.com.
Dai Sik Ko, Email: daisik.ko@gilhospital.com.
Yun Hak Kim, Email: yunhak10510@pusan.ac.kr.
Data Availability
The data that support the findings of the present study are available from the corresponding author upon reasonable request.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the National Research Foundation of Korea (NRF) funded by the Korea Government [grant number NRF-2020R1A2C1102433]; the Young Medical Scientist Research Grant through the Daewoong Foundation [grant number DY20111P]; and the Korea Medical Institute (KMI); and the Institute for Basic Science [grant number IBS-R022-D1].
Author Contribution
D.S.K., J.M.K., and Y.H.K. made substantial contributions to the conception and design. J.J.P., H.J.H., J.K., and E.J.K. collected the data and J.W.K. and Y.K. performed statistical analysis. C.-K.O., Y.L., and K.M. performed zebrafish experiments. C.-K.O. and Y.K. analysed and interpreted the data. C.-K.O. and D.S.K. wrote the manuscript. J.M.K. and Y.H.K. made critical revision.
Ethics Approval
All experiments with zebrafish were performed in accordance with the guidelines of UNIST IACUC (IACUC approval number: UNISTIACUC-15-14). All animal experiments have been performed in accordance with the ARRIVE/NC3R guidelines.
References
- 1.Kent K.C. (2014) Clinical practice. Abdominal aortic aneurysms. N. Engl. J. Med. 371, 2101–2108 10.1056/NEJMcp1401430 [DOI] [PubMed] [Google Scholar]
- 2.GBD 2017 Causes of Death Collaborators (2018) Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 392, 1736–1788 10.1016/S0140-6736(18)32203-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaikof E.L., Dalman R.L., Eskandari M.K., Jackson B.M., Lee W.A., Mansour M.A. et al. (2018) The Society for Vascular Surgery practice guidelines on the care of patients with an abdominal aortic aneurysm. J. Vasc. Surg. 67, 2.e2–77.e2 10.1016/j.jvs.2017.10.044 [DOI] [PubMed] [Google Scholar]
- 4.Powell J.T., Brady A.R., Brown L.C., Fowkes F.G., Greenhalgh R.M., Ruckley C.V. et al. (2002) Long-term outcomes of immediate repair compared with surveillance of small abdominal aortic aneurysms. N. Engl. J. Med. 346, 1445–1452 10.1056/NEJMoa013527 [DOI] [PubMed] [Google Scholar]
- 5.Kontopodis N., Pantidis D., Dedes A., Daskalakis N. and Ioannou C.V. (2016) The - Not So - Solid 5.5 cm threshold for abdominal aortic aneurysm repair: facts, misinterpretations, and future directions. Front. Surg. 3, 1 10.3389/fsurg.2016.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forsythe R.O., Newby D.E. and Robson J.M. (2016) Monitoring the biological activity of abdominal aortic aneurysms beyond ultrasound. Heart 102, 817–824 10.1136/heartjnl-2015-308779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ailawadi G., Eliason J.L. and Upchurch G.R. Jr (2003) Current concepts in the pathogenesis of abdominal aortic aneurysm. J. Vasc. Surg. 38, 584–588 10.1016/S0741-5214(03)00324-0 [DOI] [PubMed] [Google Scholar]
- 8.Kim Y.H., Lee S.J., Seo K.W., Bae J.U., Park S.Y., Kim E.K. et al. (2013) PAF enhances MMP-2 production in rat aortic VSMCs via a beta-arrestin2-dependent ERK signaling pathway. J. Lipid Res. 54, 2678–2686 10.1194/jlr.M037176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golledge J., Tsao P.S., Dalman R.L. and Norman P.E. (2008) Circulating markers of abdominal aortic aneurysm presence and progression. Circulation 118, 2382–2392 10.1161/CIRCULATIONAHA.108.802074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moris D.N. and Georgopoulos S.E. (2013) Circulating biomarkers for abdominal aortic aneurysm: what did we learn in the last decade? Int. Angiol. 32, 266–280 [PubMed] [Google Scholar]
- 11.Ioannidis J.P. (2005) Why most published research findings are false. PLoS Med. 2, e124 10.1371/journal.pmed.0020124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoon S., Tromp G., Vongpunsawad S., Ronkainen A., Juvonen T. and Kuivaniemi H. (1999) Genetic analysis of MMP3, MMP9, and PAI-1 in Finnish patients with abdominal aortic or intracranial aneurysms. Biochem. Biophys. Res. Commun. 265, 563–568 10.1006/bbrc.1999.1721 [DOI] [PubMed] [Google Scholar]
- 13.Harrison S.C., Smith A.J., Jones G.T., Swerdlow D.I., Rampuri R., Bown M.J. et al. (2013) Interleukin-6 receptor pathways in abdominal aortic aneurysm. Eur. Heart J. 34, 3707–3716 10.1093/eurheartj/ehs354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bown M.J., Jones G.T., Harrison S.C., Wright B.J., Bumpstead S., Baas A.F. et al. (2011) Abdominal aortic aneurysm is associated with a variant in low-density lipoprotein receptor-related protein 1. Am. J. Hum. Genet. 89, 619–627 10.1016/j.ajhg.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z. et al. (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawson N.D. and Weinstein B.M. (2002) In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 248, 307–318 10.1006/dbio.2002.0711 [DOI] [PubMed] [Google Scholar]
- 17.Kwon H.B., Choi Y.K., Lim J.J., Kwon S.H., Her S., Kim H.J. et al. (2012) AKAP12 regulates vascular integrity in zebrafish. Exp. Mol. Med. 44, 225–235 10.3858/emm.2012.44.3.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall C.J., Flores M.V., Davidson A.J., Crosier K.E. and Crosier P.S. (2002) Radar is required for the establishment of vascular integrity in the zebrafish. Dev. Biol. 251, 105–117 10.1006/dbio.2002.0794 [DOI] [PubMed] [Google Scholar]
- 19.Shen Y., Zhang F., Lan H., Chen K., Zhang Q., Xie G. et al. (2015) FRZB up-regulation is correlated with hepatic metastasis and poor prognosis in colon carcinoma patients with hepatic metastasis. Int. J. Clin. Exp. Pathol. 8, 4083–4090 [PMC free article] [PubMed] [Google Scholar]
- 20.Wan L., Huang J., Ni H. and Yu G. (2018) Screening key genes for abdominal aortic aneurysm based on gene expression omnibus dataset. BMC Cardiovasc. Disord. 18, 34 10.1186/s12872-018-0766-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen S., Yang D., Lei C., Li Y., Sun X., Chen M. et al. (2019) Identification of crucial genes in abdominal aortic aneurysm by WGCNA. PeerJ 7, e7873 10.7717/peerj.7873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tromp G. and Kuivaniemi H. (2009) Developments in genomics to improve understanding, diagnosis and management of aneurysms and peripheral artery disease. Eur. J. Vasc. Endovasc. Surg. 38, 676–682 10.1016/j.ejvs.2009.08.010 [DOI] [PubMed] [Google Scholar]
- 23.Wang S., Krinks M., Lin K., Luyten F.P. and Moos M. Jr (1997) Frzb, a secreted protein expressed in the Spemann organizer, binds and inhibits Wnt-8. Cell 88, 757–766 10.1016/S0092-8674(00)81922-4 [DOI] [PubMed] [Google Scholar]
- 24.Hoang B., Moos M., Vukicevic S. and Luyten F.P. (1996) Primary structure and tissue distribution of FRZB, a novel protein related to Drosophila frizzled, suggest a role in skeletal morphogenesis. J. Biol. Chem. 271, 26131–26137 10.1074/jbc.271.42.26131 [DOI] [PubMed] [Google Scholar]
- 25.Thysen S., Luyten F.P. and Lories R.J. (2015) Loss of Frzb and Sfrp1 differentially affects joint homeostasis in instability-induced osteoarthritis. Osteoarthritis Cartilage 23, 275–279 10.1016/j.joca.2014.10.010 [DOI] [PubMed] [Google Scholar]
- 26.Adachi H., Majima S., Kon S., Kobayashi T., Kajino K., Mitani H. et al. (2004) Niban gene is commonly expressed in the renal tumors: a new candidate marker for renal carcinogenesis. Oncogene 23, 3495–3500 10.1038/sj.onc.1207468 [DOI] [PubMed] [Google Scholar]
- 27.Carvalheira G., Nozima B.H. and Cerutti J.M. (2015) microRNA-106b-mediated down-regulation of C1orf24 expression induces apoptosis and suppresses invasion of thyroid cancer. Oncotarget 6, 28357–28370 10.18632/oncotarget.4947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai A., Zhou Y. and Li L. (2015) Rho-GTPase and atherosclerosis: pleiotropic effects of statins. J. Am. Heart Assoc. 4, 10.1161/JAHA.115.002113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laufs U., Marra D., Node K. and Liao J.K. (1999) 3-Hydroxy-3-methylglutaryl-CoA reductase inhibitors attenuate vascular smooth muscle proliferation by preventing rho GTPase-induced down-regulation of p27(Kip1). J. Biol. Chem. 274, 21926–21931 10.1074/jbc.274.31.21926 [DOI] [PubMed] [Google Scholar]
- 30.Maruhashi T., Noma K., Iwamoto Y., Iwamoto A., Oda N., Kajikawa M. et al. (2014) Critical role of exogenous nitric oxide in ROCK activity in vascular smooth muscle cells. PLoS ONE 9, e109017 10.1371/journal.pone.0109017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uehata M., Ishizaki T., Satoh H., Ono T., Kawahara T., Morishita T. et al. (1997) Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature 389, 990–994 10.1038/40187 [DOI] [PubMed] [Google Scholar]
- 32.Wassmann S., Laufs U., Baumer A.T., Muller K., Konkol C., Sauer H. et al. (2001) Inhibition of geranylgeranylation reduces angiotensin II-mediated free radical production in vascular smooth muscle cells: involvement of angiotensin AT1 receptor expression and Rac1 GTPase. Mol. Pharmacol. 59, 646–654 10.1124/mol.59.3.646 [DOI] [PubMed] [Google Scholar]
- 33.Sawada N., Itoh H., Ueyama K., Yamashita J., Doi K., Chun T.H. et al. (2000) Inhibition of rho-associated kinase results in suppression of neointimal formation of balloon-injured arteries. Circulation 101, 2030–2033 10.1161/01.CIR.101.17.2030 [DOI] [PubMed] [Google Scholar]
- 34.Liu H.W., Halayko A.J., Fernandes D.J., Harmon G.S., McCauley J.A., Kocieniewski P. et al. (2003) The RhoA/Rho kinase pathway regulates nuclear localization of serum response factor. Am. J. Respir. Cell Mol. Biol. 29, 39–47 10.1165/rcmb.2002-0206OC [DOI] [PubMed] [Google Scholar]
- 35.Sakalihasan N., Michel J.B., Katsargyris A., Kuivaniemi H., Defraigne J.O., Nchimi A. et al. (2018) Abdominal aortic aneurysms. Nat. Rev. Dis. Primers 4, 34 10.1038/s41572-018-0030-7 [DOI] [PubMed] [Google Scholar]
- 36.Consortium T.A. (2008) Genome Wide Association Studies: identifying the genes that determine the risk of abdominal aortic aneurysm. Eur. J. Vasc. Endovasc. Surg. 36, 395–396 10.1016/j.ejvs.2008.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones G.T., Tromp G., Kuivaniemi H., Gretarsdottir S., Baas A.F., Giusti B. et al. (2017) Meta-analysis of genome-wide association studies for abdominal aortic aneurysm identifies four new disease-specific risk loci. Circ. Res. 120, 341–353 10.1161/CIRCRESAHA.116.308765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toghill B.J., Saratzis A., Freeman P.J., Sylvius N. and Bown M.J. (2018) SMYD2 promoter DNA methylation is associated with abdominal aortic aneurysm (AAA) and SMYD2 expression in vascular smooth muscle cells. Clin. Epigenet. 10, 29 10.1186/s13148-018-0460-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lenk G.M., Tromp G., Weinsheimer S., Gatalica Z., Berguer R. and Kuivaniemi H. (2007) Whole genome expression profiling reveals a significant role for immune function in human abdominal aortic aneurysms. BMC Genomics 8, 237 10.1186/1471-2164-8-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biros E., Gabel G., Moran C.S., Schreurs C., Lindeman J.H., Walker P.J. et al. (2015) Differential gene expression in human abdominal aortic aneurysm and aortic occlusive disease. Oncotarget 6, 12984–12996 10.18632/oncotarget.3848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gabel G., Northoff B.H., Weinzierl I., Ludwig S., Hinterseher I., Wilfert W. et al. (2017) Molecular fingerprint for terminal abdominal aortic aneurysm disease. J. Am. Heart Assoc. 6, e006798 10.1161/JAHA.117.006798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boddy A.M., Lenk G.M., Lillvis J.H., Nischan J., Kyo Y. and Kuivaniemi H. (2008) Basic research studies to understand aneurysm disease. Drug News Perspect. 21, 142–148 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of the present study are available from the corresponding author upon reasonable request.