Figure 1.
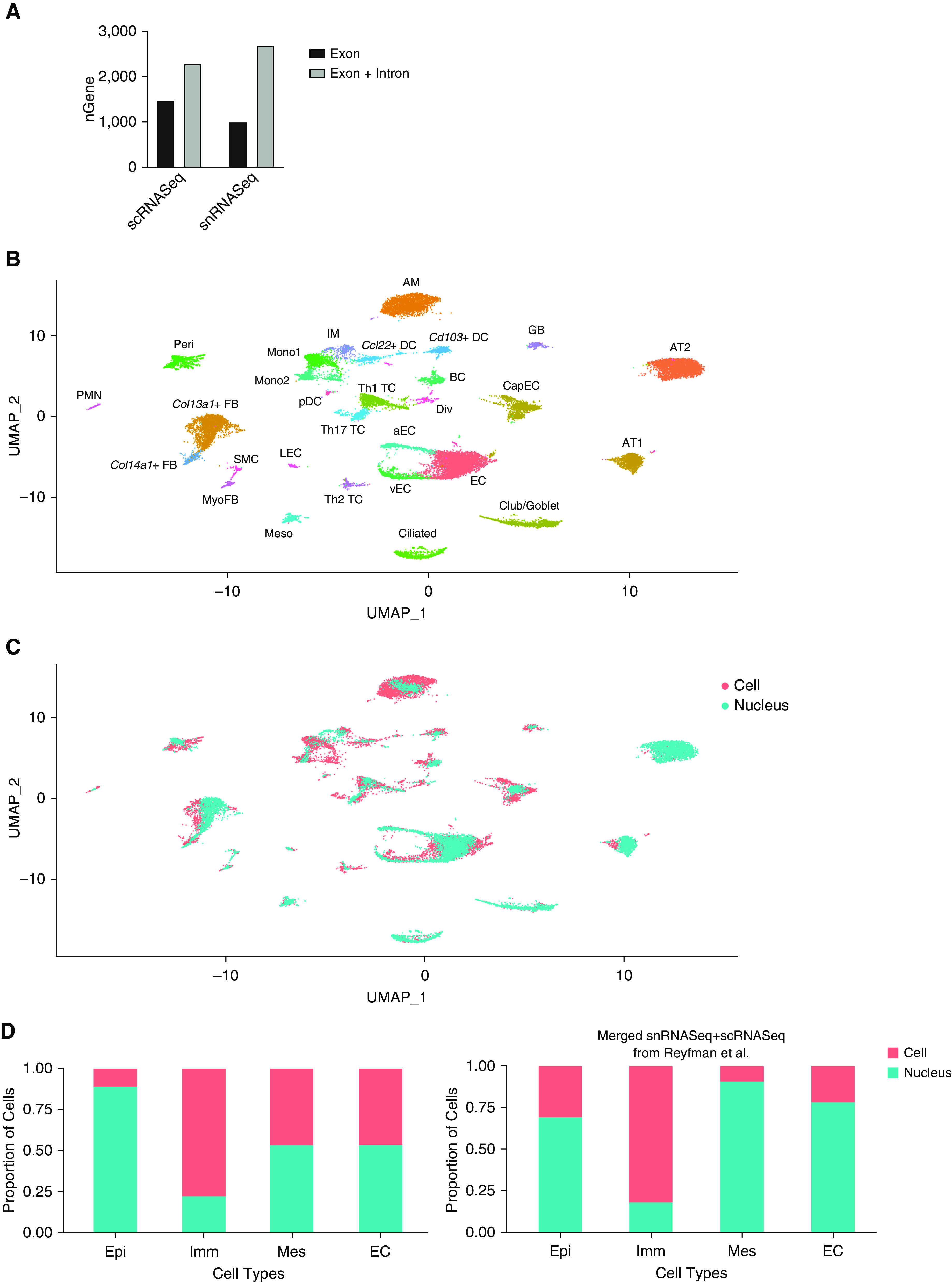
Single-nucleus RNA sequencing (snRNASeq) offers similar gene detection and improves dissociation bias compared with single-cell RNA sequencing (scRNASeq). (A) Genes detected per cell when exonic reads alone or exonic and intronic reads are used in mapping. (B) Uniform manifold approximation and projection plot of 27,978 nuclei and cells from merged snRNAseq and scRNASeq data (combined from three mice) with annotation of cell types. (C) Labeling cells from snRNASeq and scRNASeq shows minimal batch effect after Harmony. (D) Percentage contributions of snRNASeq and scRNASeq to cell categories after random downsampling to equalize numbers of cells and nuclei. Results from the present study (left) and after merge with a previously published scRNASeq dataset (right). aEC = arterial endothelial cells; AM = alveolar macrophages; AT1 = alveolar type 1 epithelial cells; AT2 = alveolar type 2 epithelial cells; BC = B cells; CapEC = capillary endothelial cells; Col13a1 = collagen type XIII α 1 chain; Col14a1 = collagen type XIV α1 chain; DC = dendritic cells; Div = dividing cells, EC = endothelial cells; Epi = epithelial; FB = fibroblasts; GB = germinal B cells, IM = interstitial macrophages; Imm = Immune; LEC = lymphatic endothelial cells; Mes = mesothelial cells; Meso = mesothelial cells; Mono = monocytes; MyoFB = myofibroblasts; NK = natural killer cells; pDC = plasmacytoid dendritic cells; Peri = pericytes; PMN = neutrophils; SMC = smooth muscle cells; TC = T cells; uMAP = uniform manifold approximation and projection; vEC = venous endothelial cells.
