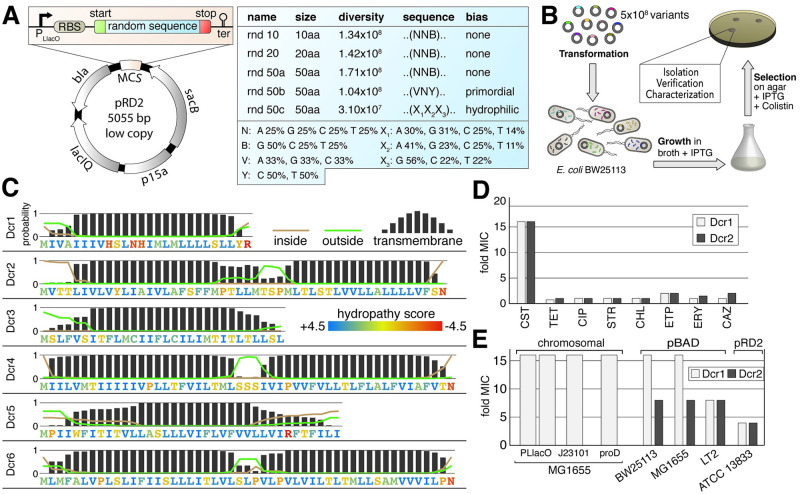
Fig 1. Selection of peptides that confer colistin resistance.
(A) Design of the expression vector and random libraries. (B) Schematic representation of the screen for inserts conferring colistin resistance. (C) Sequence analysis of the six identified de novo colistin resistance peptides (Dcr 1–6). The letters represent the amino acid sequence and are color-coded for their hydropathy score. The bars and line graphs show the probability of transmembrane domains (bars) and cytoplasmic (brown) or periplasmic (green) localization. (D) Resistance profile of E. coli BW25113 expressing Dcr1 or 2 from the IPTG-inducible pRD2 vector compared to an empty vector control strain. Minimal inhibitory concentrations (MIC) of colistin (CST) were determined by broth dilution using Sensititre microtiter plates. MICs of tetracycline (TET), ciprofloxacin (CIP), streptomycin (STR), chloramphenicol (CHL), ertapenem (ETP), erythromycin (ERY) and ceftazidime (CAZ) were determined using Etest on agar plates. (E) MICs of colistin for various constructs expressing Dcr1 or 2 compared to empty vector control strains. All MIC determinations were performed at least in triplicate.