SUMMARY
The telomeric shelterin protein telomeric repeat-binding factor 2 (TRF2) recruits origin recognition complex (ORC) proteins, the foundational building blocks of DNA replication origins, to telomeres. We seek to determine whether TRF2-recruited ORC proteins give rise to functional origins in telomere repeat tracts. We find that reduction of telomeric recruitment of ORC2 by expression of an ORC interaction-defective TRF2 mutant significantly reduces telomeric initiation events in human cells. This reduction in initiation events is accompanied by telomere repeat loss, telomere aberrations and dysfunction. We demonstrate that telomeric origins are activated by induced replication stress to provide a key rescue mechanism for completing compromised telomere replication. Importantly, our studies also indicate that the chromatin remodeler SNF2H promotes telomeric initiation events by providing access for ORC2. Collectively, our findings reveal that active recruitment of ORC by TRF2 leads to formation of functional origins, providing an important mechanism for avoiding telomere dysfunction and rescuing challenged telomere replication.
Graphical Abstract
In Brief
Drosopoulos et al. report that active recruitment of ORC2 to telomeres by TRF2 leads to formation of functional DNA replication origins in human telomeres. These origins provide an important mechanism for avoiding telomere repeat loss and dysfunction and rescuing challenged telomere replication.
INTRODUCTION
Genomic instability resulting from telomere dysfunction is an important contributing factor to cancer as well as congenital and age-related degenerative diseases (Blackburn et al., 2015; Martínez and Blasco, 2017). Maintenance of proper telomere structure and function is dependent on efficient replication of telomeric DNA. Telomeres challenge the replication machinery, in part because of the secondary structure-prone nature of their G-rich repetitive DNA sequence, which can stall replication forks (Anand et al., 2012; Ivessa et al., 2002; Makovets et al., 2004; Miller et al., 2006; Sfeir et al., 2009; Verdun and Karlseder, 2006), potentially leading to incomplete replication and dysfunction. Among the mechanisms employed to rescue stalled replication is to initiate replication ahead of the stall so that a converging fork from the opposite direction completes replication of the DNA beyond the stalled fork. Because telomeres are heavily heterochromatinized (Blasco, 2007), they are predicted to have closed chromatin repressive to initiation. This implies that telomeric origins would be unavailable for resuming stalled telomere replication. However, we have shown that, in fact, replication can initiate within the telomere (Drosopoulos et al., 2012, 2015), indicating that telomeres are not inherently restrictive to initiation and that telomeric origins could potentially be used if needed to rescue stalled replication.
DNA replication initiation in eukaryotic cells is mediated by the origin recognition complex (ORC) (Hyrien, 2015; Urban et al., 2015). In mammalian cells, binding of ORC proteins to genomic DNA does not generally seem to involve defined sequences (Hyrien, 2015; Urban et al., 2015). Instead, chromatin status appears to be a key determinant of ORC binding. Mapping of ~52,000 ORC2 genomic binding sites in human cells revealed that the ORC binds without sequence specificity to open (DNase I-hypersensitive) regions (Miotto et al., 2016). The extensive heterochromatization of telomeres suggests that telomeres are relatively inaccessible to ORC proteins and that active mechanisms exist that allow ORC recruitment and subsequent formation of telomeric origins. Indeed, we and others have demonstrated that the telomere-specific shelterin protein TRF2 can interact with and recruit ORC proteins to telomeres (Deng et al., 2007, 2009; Higa et al., 2017; Tatsumi et al., 2008). However, it has yet to be established whether the recruited ORC proteins give rise to functional replication origins.
The current paradigm is that telomere replication is critically dependent on prevention of stalling/collapse of the replication fork that is copying the telomere. This is based on the perceived inability to restart replication ahead of a collapsed fork by activating telomeric origins (Ahmed and Lingner, 2018; Gilson and Géli, 2007; Oganesian and Karlseder, 2009). However, TRF2 recruitment of ORC likely results in many potential origins in telomeres (Atanasiu et al., 2006; Deng et al., 2007; Tatsumi et al., 2008). Furthermore, the fact that telomeres are genomic loci where ORC proteins are actively and specifically recruited implies a key role of telomeric origins in telomere replication. Therefore, we investigated whether ORC protein recruitment by TRF2 can lead to formation of active replication origins. We have demonstrated previously that expression of an ORC interaction-defective mutant of TRF2 in human cells reduces telomeric recruitment of ORC2 (Deng et al., 2007). We now show that reduced TRF2-mediated telomeric recruitment of ORC2 significantly reduces telomeric initiation events, accompanied by telomere repeat loss and dysfunction. We also find that telomeric origins can be activated in response to replication stress to provide a rescue mechanism for completion of telomere replication. Thus, in addition to prevention of fork stalling, our studies reveal that activation of telomeric origins is a key mechanism by which cells avoid telomere dysfunction.
RESULTS
Telomere Replication Predominantly Initiates within the Ch7q Telomere in Human LOX Cells
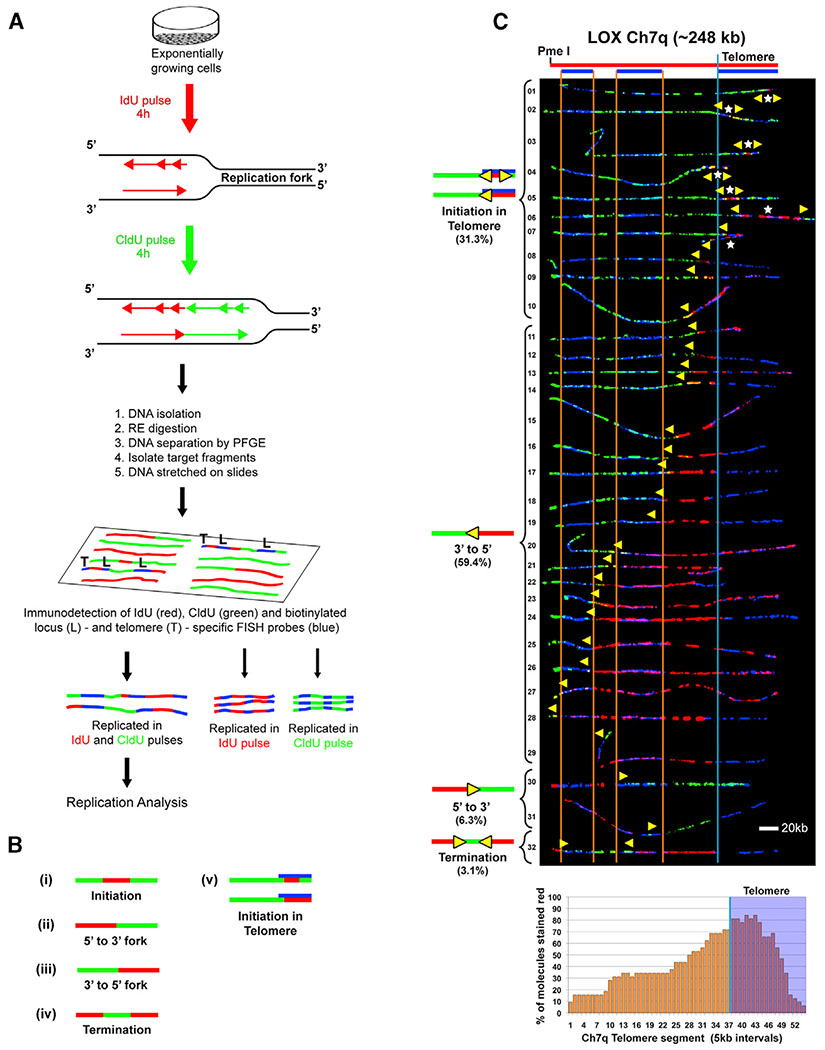
Our previous studies established that human telomere replication most frequently initiates outside of the telomere, although we did detect telomeric origins (Drosopoulos et al., 2012). However, initiation events detected by single-molecule analysis of replicated DNA (SMARD), the DNA fiber-based replication analysis we employed (described below), are generally seen within stretches of replicated DNA spanning at least 5 kb. Therefore, some telomeric origins may have gone undetected because of the relatively short average length (~8–10 kb) of the human telomeres we studied. Furthermore, we found that telomere length influenced the frequency of telomere initiation, where increased telomere length resulted in more detected telomeric initiation events (Drosopoulos et al., 2012). This prompted us to examine telomere origin use in LOX human melanoma cells (Fodstad et al., 1988), which have extremely long telomeres (>40 kb on average) compared to most human cells (5–15 kb on average; Samassekou et al., 2010). This long telomere length provides a larger region for initiation events to occur within the telomere as well as to be visualized. Despite their relatively long length, telomeres in LOX cells are maintained like in most other cell lines, via a telomerase-based mechanism (Mahalingam et al., 2011; Nakamura et al., 1997). We selected a 250-kb genomic segment from the end of chromosome arm 7q (Ch7q), containing the telomere and associated subtelomeric region, as a model locus. This segment was selected because it was anticipated that telomeric origins would be used frequently to replicate the telomere. We observed previously that this (Ch7q) segment is replicated with similar frequency by forks originating within (or near) the telomere or outside of the segment (Drosopoulos et al., 2012). We examined telomere replication using SMARD (Drosopoulos et al., 2012; Norio and Schildkraut, 2001). In SMARD, asynchronous cell cultures are pulse-labeled sequentially with two distinguishable nucleoside analogs, iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU). Individual molecules of pulse-labeled replicated DNA are isolated and immunostained, and labeling patterns are revealed by fluorescence microscopy imaging. Regions replicated during the first (IdU) pulse appear red, and regions replicated during the second (CldU) pulse appear green (Figure 1A). Specific labeled telomere-containing molecules of interest are identified and oriented by asymmetric signal patterns (blue bar codes) produced by locus-specific fluorescence in situ hybridization (FISH) probes and a telomere-specific peptide nucleic acid (PNA) probe. The labeling patterns reveal and map the position, direction, and density of the replication forks in the population of replicated molecules (Norio and Schildkraut, 2001). From this information, the sites of replication initiation and termination in a given genomic region can be established (Figure 1B).
Figure 1. Telomere Replication Predominantly Initiates within the Ch7q Telomere in Human LOX Cells.
(A) SMARD. Replicating DNA in exponentially and asynchronously growing cells is labeled sequentially by pulsing with IdU for 4 h, followed by CldU for 4 h. Labeled cells are embedded in agarose, lysed, and deproteinized to release genomic DNA. The genomic DNA is digested with a rare-cutting restriction endonuclease to obtain unique large molecules, which are separated by pulsed-field gel electrophoresis (PFGE) to enrich for by size for the particular segments of interest. PCR screening is performed to identify the target fragment within the gel, which is excised. The gel slice containing the segment of interest is melted, and the enriched DNA in the melted gel solution is stretched on silanized glass slides. Indirect immunofluorescence with antibodies against the halogenated nucleosides is used to identify regions where IdU (red) or CldU (green) was incorporated into the DNA. Immunodetected biotinylated probes (blue) are used to identify the molecules to be analyzed (Drosopoulos et al., 2012). Images of molecules fully labeled with IdU, CldU, or both IdU and CldU and that also display all probes are collected. Molecules fully labeled with red and green (IdU and CldU) are used for replication analysis, whereas fully red (IdU) or fully green (CldU) molecules serve as labeling controls.
(B) Staining patterns of molecules indicate the direction of replication fork progression because replication in IdU occurs first. Molecules where one side is stained red and the other side is stained green indicate that forks progressed in a single direction through the segment (ii and iii). Initiation events are observed as a red tract flanked on both sides by green (i), whereas a green tract flanked on both sides by red indicates a termination event (iv). In addition to a red tract flanked on both sides by green, initiation events in the telomere (v) can appear as a red tract that does not extend out of the telomere or extends only a few kilobases out from the telomere.
(C) SMARD analysis of a segment of Ch7q containing the telomere from LOX cells. Alignments of replicated molecules fully labeled with IdU (red) and CldU (green) are shown, collected from four independent samples stretched on slides (156 fully red- and 164 fully green-labeled molecules were also collected). A map of the 7q locus is depicted above the alignment, with the positions of the FISH probes (blue bars below) used to identify and orient the molecules indicated. Vertical orange lines mark positions of the ends of the subtelomeric FISH signals used to align the molecules. The boundary between the subtelomere and telomere is delineated by a vertical blue line. Yellow arrows mark sites of transition from IdU to CldU incorporation and indicate the direction of fork progression at the moment of transition during replication (polarity of fork direction assigned relative to the polarity of the telomere G-rich strand of the segment). Initiation events (red tracts flanked by green) are seen in telomeres. White stars indicate the centers of initiations. A replication profile, a histogram of the percentages of molecules containing red staining (IdU) per 5-kb interval along the segment, is shown under the alignment. The 5-kb intervals that have the highest percentage of molecules stained red are those that, on average, replicate first. The telomere portion of the segment is shaded in blue, and the telomere-subtelomere boundary is indicated by a blue vertical line.
SMARD analysis of the LOX Ch7q locus revealed a striking feature of telomere replication at the locus. Approximately one third of the molecules examined displayed telomeric initiation events. Most (70%) of these events were observed as a red tract flanked by green tracts (Figure 1C). The remaining initiation events were detected as a red tract that does not extend beyond the telomere or extends only a few kilobases beyond the telomere/subtelomere junction (Figure 1C molecules 7–10), indicating that origins that gave rise to these forks must have been located within the telomere. In addition, almost all (~94%) of the Ch7q molecules displayed continuous red labeling that extends from the telomere into the subtelomere, indicating the presence of an origin within the red-labeled region. By plotting the percentage of molecules containing IdU (first label) incorporation (red staining) for each 5 kb of the DNA segment, a replication profile–a graphical summary of replication order (timing) within the segment–is obtained (Figure S1). The 5-kb intervals that have the highest percentage of molecules stained red are those that, on average, replicate first. As indicated by the replication profile for Ch7q, the percentage of red staining is highest in the telomere (Figure 1C); thus, the telomere is replicated earlier than the subtelomere. Because no subtelomeric origins were observed compared with 10 observed telomeric initiations (Figure 1C), this strongly suggests that telomere replication in molecules with continuous red labeling from the right (telomere) end of the segment was likely initiated within the telomere. These findings indicate that the Ch7q telomere appears to be replicated by forks originating from initiation events within the telomere. Thus, the LOX Ch7q telomere is the first human telomere identified that appears to be replicated primarily from telomeric origins.
Telomere Replication Occasionally Initiates within the Ch10q Telomere in LOX Cells
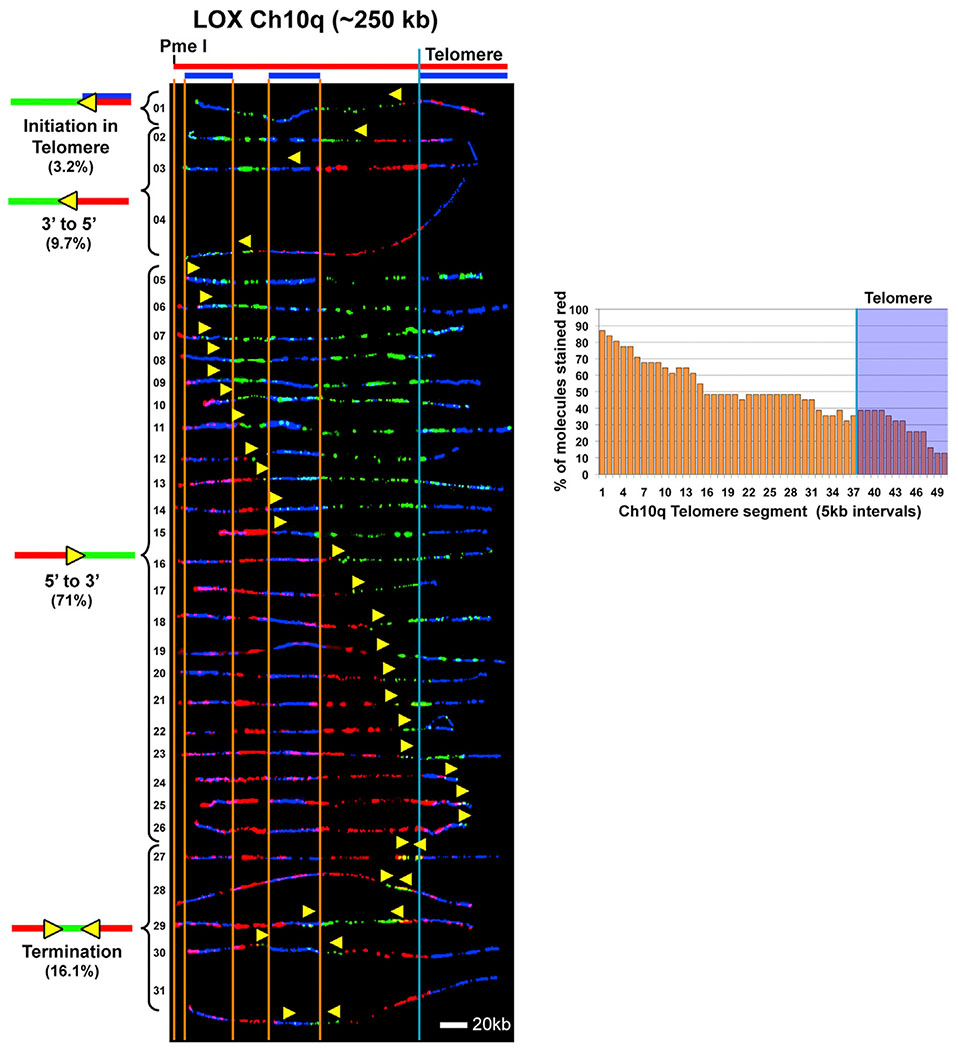
The extensive use of telomeric origins we detected in long LOX Ch7q telomeres was consistent with our previous observations showing that increasing telomere length results in more frequent telomeric initiations (Drosopoulos et al., 2012). This supported the idea that telomere length is a key determinant of telomeric origin use and that origins fire more often in longer telomeres. It is then anticipated that LOX cells should, in general, frequently use telomeric origins to replicate their long (45 kb on average) telomeres. To test whether this is the case, we next examined replication of the chromosome 10q telomere locus. This locus was chosen because we found previously that it showed few telomeric initiations in cells with average-length (5–12 kb) telomeres but had increased telomeric origin use in cells with somewhat longer (20–25 kb) telomeres (Drosopoulos et al., 2012). We examined replication of a 250-kb genomic segment from the end of the 10q chromosome arm (Ch10q) containing the telomere and associated subtelomeric region. Our SMARD analysis revealed that, unexpectedly, most (71%) of the molecules showed no evidence that replication began in the telomere (Figure 2). Of the remaining (29%) molecules, less than half (4 of 9) showed evidence that replication likely initiated in the telomere (Figure 2, molecules 1 and 27–29). Furthermore, the replication profile of Ch10q (Figure 2) shows that the subtelomere is generally replicated before the telomere. These data indicate that the LOX Ch10q telomere is replicated primarily from origins outside of the telomere. Significantly, these findings indicate that long telomere length is not always accompanied by frequent use of telomeric origins and that increased telomere length does not always promote initiation in the telomere.
Figure 2. Telomere Replication Occasionally Initiates within the Ch10q Telomere in LOX Cells.
SMARD analysis of a segment of Ch10q containing the telomere from LOX cells. Alignments of replicated molecules fully labeled with IdU (red) and CldU (green) are shown, collected from four independent samples stretched on slides (173 fully red- and 147 fully green-labeled molecules were also collected). Vertical lines (orange and blue) demarcate the boundaries where FISH probes bind, as described in Figure 1. Symbols are as in Figure 1. A replication profile histogram is shown to the right of the molecule alignment.
Expression of TRF2ΔB Decreases Replication Initiation within Telomeres in LOX Cells
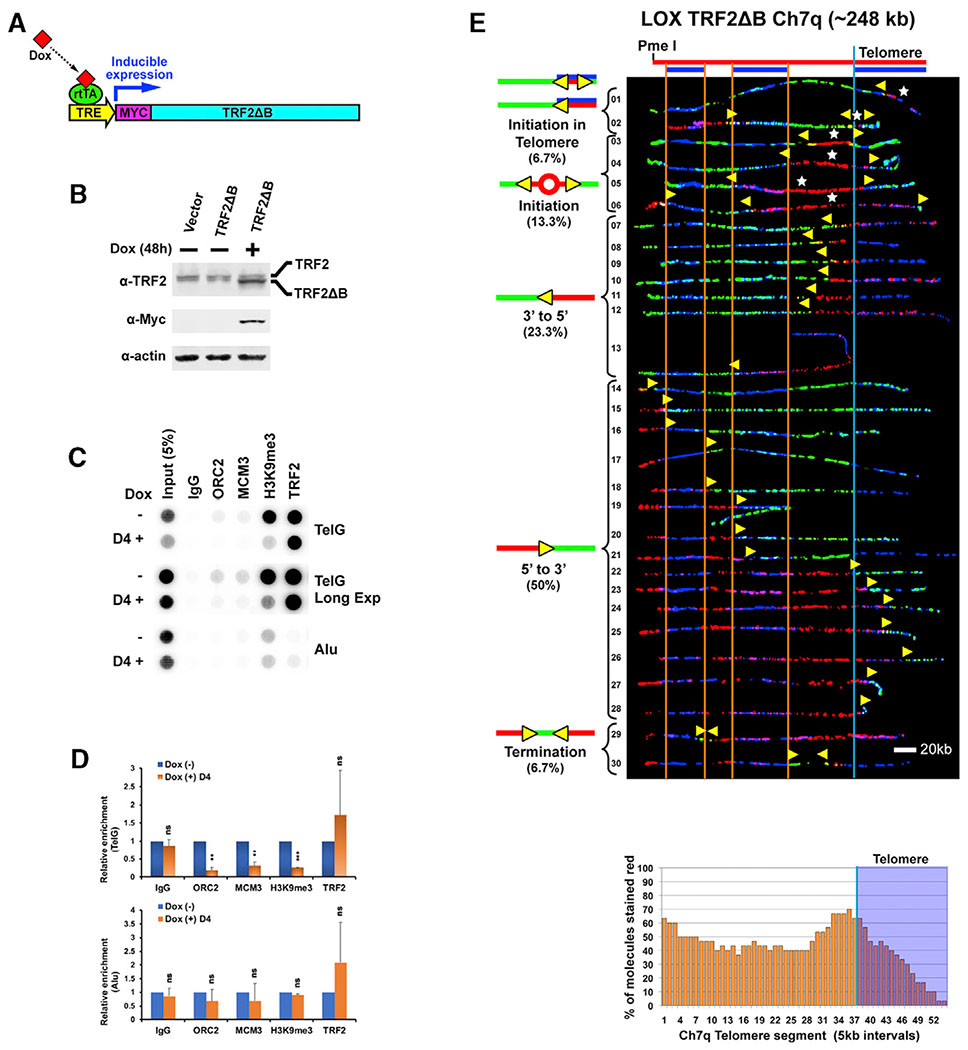
To understand how initiation in the telomere is accomplished, we investigated whether TRF2 has a role in this process. Work from our group and others has shown that TRF2 recruits ORC proteins, the building blocks of origins, to telomeres (Atanasiu et al., 2006; Deng et al., 2007; Higa et al., 2017; Tatsumi et al., 2008). We have found that recruitment of ORC2 is mediated by interactions between ORC2 and the N-terminal basic (B) domain of TRF2 (Deng et al., 2007). We have shown that TRF2ΔB, a telomere-binding TRF2 deletion mutant lacking the B domain (Wang et al., 2004), is ORC interaction defective and that ectopic expression of TRF2ΔB reduces telomeric recruitment of ORC2 (Deng et al., 2007). Accordingly, we reasoned that disruption of ORC telomeric recruitment by TRF2ΔB expression should result in reduced telomeric origin formation and use. Thus, we examined the effect of TRF2ΔB expression on telomeric initiation events to determine whether ORC protein recruitment by TRF2 gives rise to active telomeric origins. We first generated a TRF2ΔB-expressing LOX cell line, LOX TRF2ΔB, where TRF2ΔB expression is from a stably integrated construct, under control of a doxycycline-inducible promoter (Figures 3A and 3B). To determine whether TRF2ΔB expression in these cells leads to reduced telomeric recruitment of ORC, the cells were analyzed by chromatin immunoprecipitation (ChIP) assay. We found that doxycycline-induced expression of TRF2ΔB reduced ORC2 association with telomeric DNA by ~82% (Figures 3C and 3D). In addition, disruption of ORC recruitment reduced the amount of telomere-bound MCM3, a pre-replication complex (pre-RC) protein, by ~69%, indicating a decrease in potentially available, licensed telomeric origins (Figures 3C and 3D). In comparison, expression of full-length TRF2 resulted in an ~20% reduction in ORC2 association with telomeric DNA and no change in telomere-bound MCM3 (Figures S2A, S2B, and S2D). We also found that disruption of ORC recruitment by TRF2ΔB reduced the amount of telomere-bound H3K9me3 histone by ~74% (Figures 3C and 3D). We have found previously that the ORC is essential for establishing telomeric heterochromatin (Deng et al., 2009). Thus, this reduction in H3K9me3 may be correlated with changes in telomeric heterochromatin resulting from the reduction in ORC recruitment after TRF2ΔB expression.
Figure 3. Expression of TRF2ΔB Reduces ORC2 Recruitment to LOX Cell Telomeres and Decreases Replication Initiation within the Ch7q Telomere in LOX Cells.
(A) Doxycycline (Dox)-inducible expression of Myc-tagged TRF2ΔB. rtTA, reverse tetracycline trans-activator; TRE, tetracycline response element.
(B) Immunoblot of cell lysates of LOX cell line stably expressing Dox-inducible TRF2ΔB. Untagged endogenous WT TRF2 (upper band) and Myc-tagged TRF2ΔB (lower band) are indicated.
(C) ChIP analysis of LOX TRF2ΔB cells grown in the absence (−) or presence (+) of 1 μg/mL Dox for 4 days using antibodies against TRF2, ORC2, MCM3, H3K9me3, or immunoglobulin G (IgG) (control). Blots were probed by hybridization with 32P-labeled telomeric TTAGGG (TelG) or Alu repeat (Alu) probes. Long Exp, long exposure.
(D) Quantification of at least three independent experiments represented in (C). The ChIP data were first normalized to input, and the percent input for Dox (+) is shown as relative to dox (−), which was set as 1. Student’s t test was used for statistical analysis. Error bars indicate SD. **p < 0.01, ***p < 0.001; ns, no statistical significance (p > 0.05).
(E) SMARD analysis of the Ch7q telomere segment from LOX TRF2ΔB cells grown for 3 days in the presence of 1 μg/mL Dox. Alignments of replicated molecules fully labeled with IdU (red) and CldU (green) are shown, collected from six independent samples stretched on slides (76 fully red- and 79 fully green-labeled molecules were also collected). Vertical lines (orange and blue) demarcate the boundaries where FISH probes bind, as described in Figure 1. Symbols are as in Figure 1. A replication profile histogram is shown under the molecule alignment.
We next analyzed replication progression through the Ch7q telomere locus in TRF2ΔB-expressing cells by SMARD to determine whether a reduction in TRF2-mediated recruitment of the ORC leads to fewer telomeric origins. We found a marked reduction in telomeric initiation events and telomere-originated forks in TRF2ΔB-expressing cells compared with cells expressing wild-type (WT) TRF2. The majority (60%) of LOX TRF2ΔB telomere-containing molecules showed no evidence that replication began in the telomere (Figure 3E, molecules 3–5 and 14–28). In comparison, only ~6% of molecules from LOX cells (expressing WT TRF2) lacked evidence of telomeric initiation (Figure 1C, molecules 30 and 31). Of the remaining 40% of LOX TRF2ΔB molecules, most (9 of 12) showed evidence that replication likely initiated in the telomere (Figure 3E, molecules 1, 2, 7–12, and 30). SMARD analysis was also performed on empty vector-containing LOX cells treated with doxycycline for 3 days as a control to account for any effects of doxycycline treatment during TRF2ΔB induction. Replication of the Ch7q segment in these cells (Figure S3A) was identical to replication in LOX cells (Figure 1). To confirm the effect of TRF2ΔB expression on telomere origin use, we also examined telomeric initiation in the Ch10q locus. Similar to the Ch7q locus, we found a distinct reduction in telomeric initiation events and telomere-originated forks in TRF2ΔB-expressing cells. Only ~3% of LOX TRF2ΔB Ch10q molecules (1 of 32) showed evidence that replication likely initiated in the telomere (Figure S4, molecule 32). In comparison, ~13% of molecules from LOX cells (expressing WT TRF2) displayed evidence of telomeric initiation (Figure 2, molecules 1 and 27–29). Replication of the Ch10q segment in control empty vector-containing LOX cells treated with doxycycline (Figure S3B) was identical to replication in LOX cells (Figure 2). Collectively, these results indicate that active recruitment of ORC, mediated by TRF2, leads to functional origins and replication initiation in telomeres.
Expression of TRF2ΔB Results in Telomere Repeat Loss and Dysfunction in LOX Cells
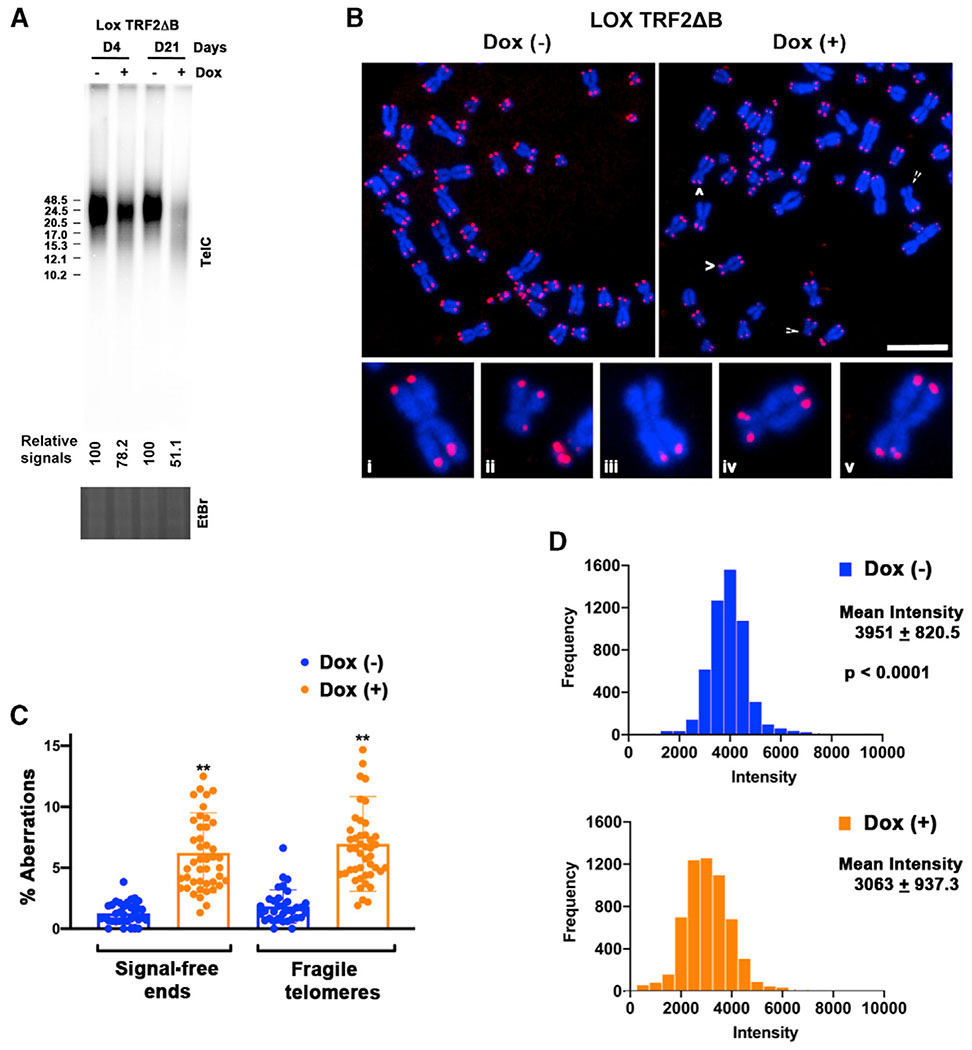
One potential outcome of telomeric initiation failing to occur is that telomeres are put at increased risk of incomplete replication and shortening, leading to dysfunction. Therefore, we determined whether the reduced telomeric initiation seen in LOX TRF2ΔB cells was accompanied by telomere loss, fragility (a manifestation of incompletely replicated regions), and dysfunction. We found that total telomere signal intensity in LOX cells, measured by telomere restriction fragment length assay (TLA), was reduced by ~22% after 4 days and ~49% after 21 days of continuous TRF2ΔB expression compared with controls (Figure 4A). In addition, telomere shortening was observed in TRF2ΔB-expressing cells at 21 days in the TLAs (Figure 4A). In comparison, expression of full-length TRF2 for 4 days resulted in no change in telomere signal intensity and a slight change after 10 days (Figure S2C). We then examined the effect of TRF2ΔB expression on metaphase telomeres, including induction of fragile telomeres, by telomere FISH. Fragile telomeres are abnormal telomeric structures observed on metaphase chromosomes by FISH, where the telomere appears decondensed and gapped so that it is seen as multiple split foci rather than a single focus. We observed an ~4 fold increase in fragile telomeres as well as an ~5 fold increase in telomere signal-free ends after extended TRF2ΔB expression in LOX cells (Figures 4B and 4C). Overall, we observed an ~23% reduction in average telomeric signal intensity in day 21 metaphase resulting from TRF2ΔB expression (Figure 4D). These data indicate that TRF2ΔB expression in LOX cells results in telomere repeat loss and dysfunction.
Figure 4. Expression of TRF2ΔB Results in Telomere Repeat Loss and Dysfunction in LOX Cells.
(A) Telomere length analysis of LOX TRF2ΔB cells grown in the absence (−) or presence (+) of 1 μg/mL Dox for 4 or 21 days. Telomere length and relative amount of telomeric DNA were determined by restriction digestion of genomic DNA with AluI/MboI, followed by PFGE and Southern hybridization with a 32P-labeled (CCCTAA)4 probe. Ethidium bromide staining of the total DNA digest was used to normalize for DNA loading (bottom). Fragment size (in kilobases) is indicated on the left of the blot. The relative intensity of telomeric DNA signals (− Dox versus + Dox) is indicated below the blot.
(B) Representative telomere FISH analysis on metaphase spreads of LOX TRF2ΔB cells grown for 21 days in the absence (−) or presence (+) of 1 μg/mL Dox. White arrows indicate common telomere aberrations. Also shown is a magnified view of common telomere aberrations found in TRF2ΔB-expressing cells: normal telomere ends (i), telomere signal-free ends (ii–iii), and fragile telomeres (iv–v).
(C) Quantification of the percentage of observed telomeric aberrations per chromosome end in LOX TRF2ΔB cells grown for 21 days in the absence (−) or presence (+) of 1 μg/mL Dox. A total of 2,583 Dox (−) and 2,953 dox (+) metaphases were scored. Data shown were obtained from at least three independent experiments. Student’s t test was used for statistical analysis. Error bars indicate SD. **p < 0.001.
(D) Quantification of metaphase telomere FISH signal intensity in LOX TRF2ΔB cells grown for 21 days in the absence (−) or presence (+) of 1 μg/mL Dox. A total of 5,295 Dox (−) and 5,775 Dox (+) telomere ends were scored. Data shown were obtained from at least three independent experiments. Mann-Whitney test was used for statistical analysis.
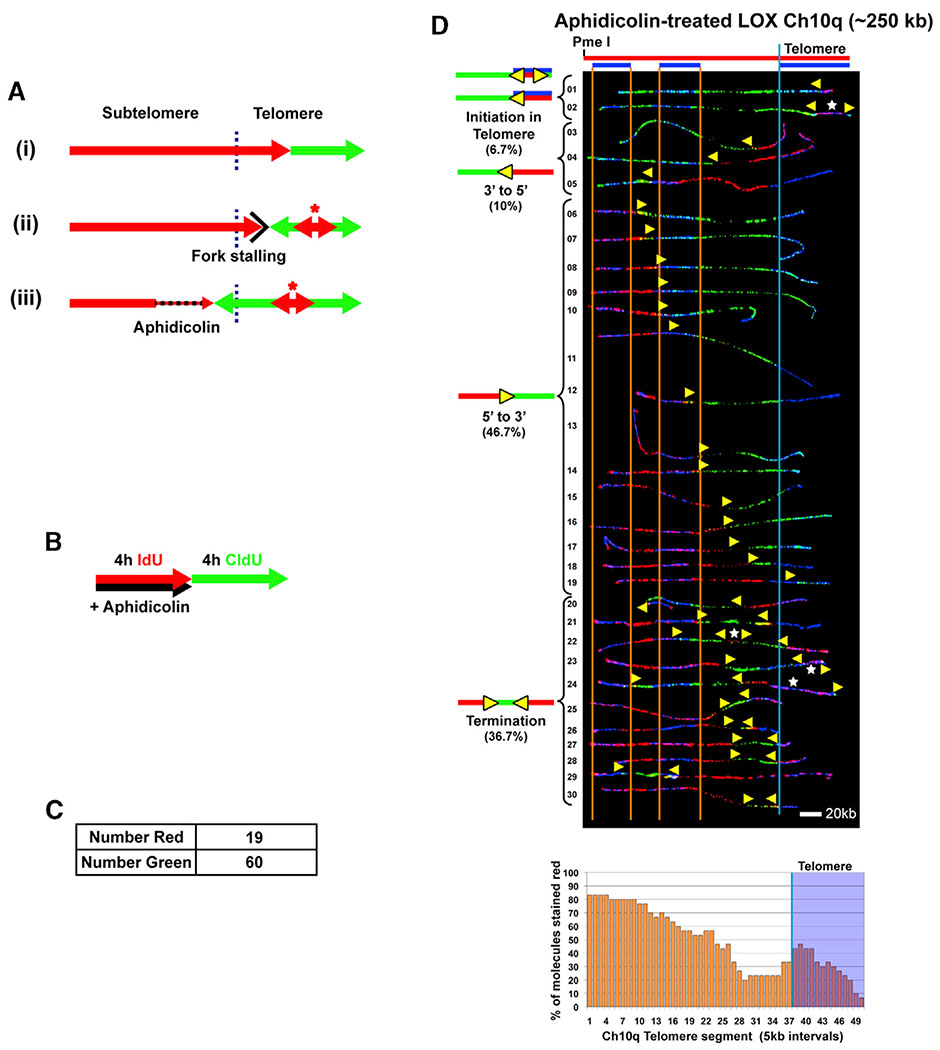
Slowdown of Telomeric Replication Forks by Aphidicolin Leads to Dormant Origin Activation in the Telomere to Rescue Slowed/Stalled Replication
In addition to functioning as DNA synthesis initiation sites during normal genome replication, origins can be activated distal to a challenged fork as a key replication stress recovery response (Ge et al., 2007; Woodward et al., 2006). Telomeric DNA challenges fork movement because of the structure-prone nature of the G-rich sequence, which can pause or stall replication forks (Anand et al., 2012; Ivessa et al., 2002; Makovets et al., 2004; Miller et al., 2006; Sfeir et al., 2009; Verdun and Karlseder, 2006). Thus, replication restart from a telomeric origin ahead of a stalled telomeric fork is a potential option to rescue the stressed fork (Figure 5A). Accordingly, this predicts that perturbed movement of forks originating from the subtelomere that are used to replicate a telomere, which is the predominant case for the Ch10q locus (Figure 2), should trigger telomeric origin firing ahead of the fork. We tested this idea by further challenging replication forks moving into/through the Ch10q telomere by low-level aphidicolin treatment (Figure 5A).
Figure 5. Slowdown of Telomeric Replication Forks by Aphidicolin Leads to Dormant Origin Activation in the Telomere to Rescue Slowed/Stalled Replication.
(A) Telomere replication by forks originating in the subtelomere (e.g., Ch10q) is normally completed without telomeric origins (i). Should replication stall/pause in the telomere, initiation within the telomere (asterisk) will allow complete replication (ii). Slowing fork progression by aphidicolin treatment should lead to telomeric origins firing to rescue slowed/stalled replication (iii). The telomere/subtelomere junction is indicated by a vertical blue line.
(B–D) SMARD analysis of the Ch10q telomere segment from LOX cells treated with aphidicolin. (B) Cells were pulse-labeled with IdU in the presence of aphidicolin (110 nM), followed by labeling with CldU in the absence of aphidicolin.
(C) Shown are the numbers of fully red stained (NR) and green stained (NG) molecules collected. The ratio of red to green molecules (19/60) indicates that replication in the presence of aphidicolin proceeded at ~1/3 of the rate as in the absence of aphidicolin.
(D) Alignments of replicated molecules fully labeled with IdU (red) and CldU (green) are shown, collected from two independent samples stretched on slides. Vertical lines (orange and blue) demarcate the boundaries where FISH probes bind, as described in Figure 1. Symbols are as in Figure 1. A replication profile histogram is shown under the molecule alignment.
We performed SMARD on LOX cells treated with aphidicolin (110 nM) during the IdU labeling pulse, followed by CldU pulsing in the absence of aphidicolin (Figure 5B). Because recovery from aphidicolin inhibition is rapid (Levenson and Hamlin, 1993), inhibitory effects on fork movement were mainly restricted to the IdU pulse. We determined relative fork speeds during each pulse by comparing the total numbers of fully IdU-labeled (red) and fully CldU-labeled (green) Ch10q molecules that were obtained (in addition to dual-labeled molecules) (Figure 5C). We observed ~1/3 as many fully red molecules as fully green molecules, indicating that replication in the presence of aphidicolin proceeded at ~1/3 of the rate as in the absence of aphidicolin. When we examined telomeric origin use, we found a 3-fold increase in telomeric initiation events in Ch10q molecules from aphidicolin-treated cells compared with untreated cells. 37% (11 of 30) of the molecules from aphidicolin-treated cells showed evidence that replication likely initiated in the telomere (Figure 5D, molecules 1,2, 21–28, and 30). In comparison, only ~13% of molecules from untreated LOX cells displayed evidence of telomeric initiation (Figure 2, molecules 1 and 27–29). Significantly, this indicated that telomeric origins can fire to provide a rescue mechanism for completion of telomere replication.
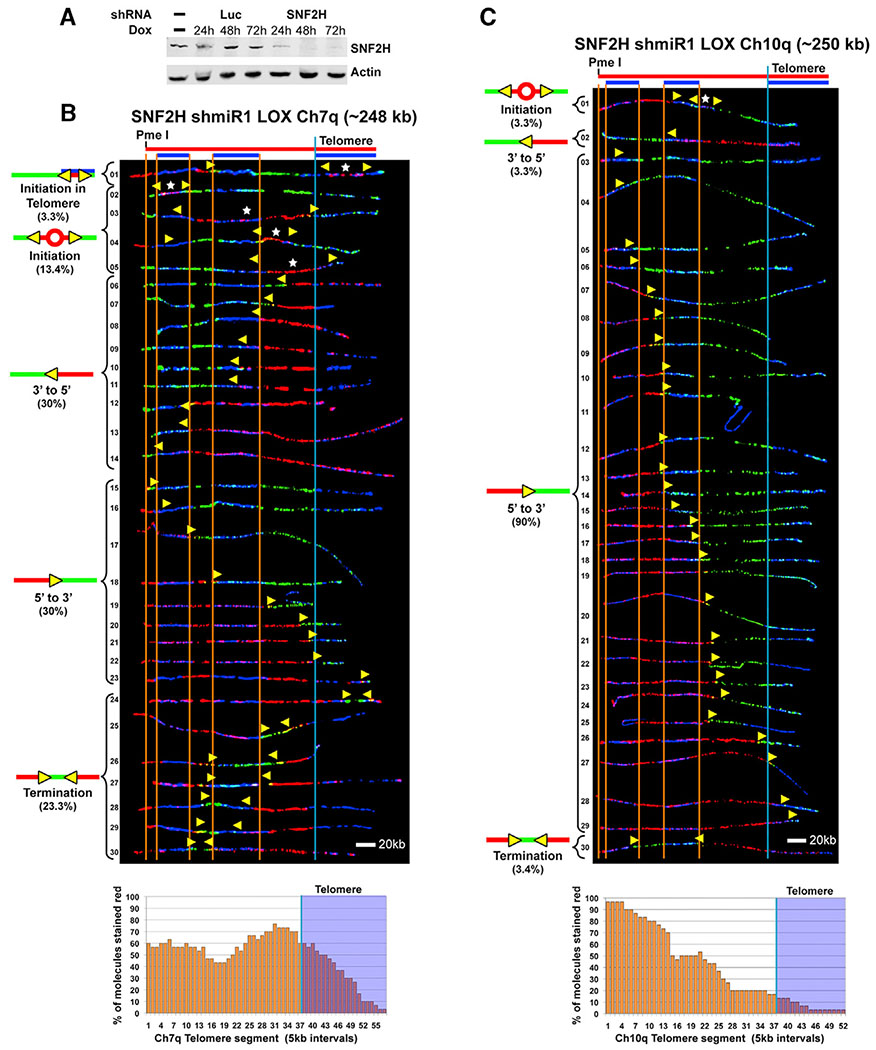
Knockdown of SNF2H Decreases Replication Initiation within Telomeres in LOX Cells
Telomeres are heavily heterochromatinized (Blasco, 2007), predicting a compact and difficult-to-access closed structure. Accordingly, we speculated that telomeric chromatin needs to be remodeled to create open regions for initiation to occur. We tested this idea by determining whether the chromatin remodeler SNF2H, which is associated with heterochromatin replication (Collins et al., 2002), assists with telomeric origin formation. Specifically, we depleted LOX cells of SNF2H and examined the cells for effects on telomeric origin activation. We used an inducible shRNA system to generate stable LOX lines expressing SNF2H-specific or luciferase (Luc) control shRNAs. A time course induction confirmed efficient depletion of SNF2H by the SNF2H-targeting shRNA but not by the control shRNA (Figure 6A). We then examined telomere replication in SNF2H-depleted LOX cells by SMARD. We observed a distinct reduction in telomeric initiation events and telomere-originated forks in SNF2H-depleted cells. 43% (13 of 30) of the SNF2H-depleted Ch7q molecules showed no evidence that replication began in the telomere ((Figure 6B, molecules 2–5 and 15–23). Of the remaining molecules, only ~10% (3 of 30) showed evidence that replication likely initiated in the telomere (Figure 6B, molecules 1, 24, and 25). In comparison, all of the control shRNA Ch7q molecules showed evidence that replication could have started in the telomere (Figure S5A), which was similar to the parental LOX cells, where ~94% of Ch7q molecules displayed evidence of telomeric initiation (Figure 1, molecules 1–29 and 32). To confirm the effect of SNF2H depletion on telomere origin use, we also examined telomeric initiation in the Ch10q locus. Similar to the Ch7q locus, we found a distinct reduction in telomeric initiation events and telomere-originated forks in SNF2H depleted cells. Only ~7% of LOX SNF2H-depleted Ch10q molecules (2 of 30) showed evidence that replication could have initiated in the telomere (Figure 6C, molecules 2 and 30). In comparison, ~26% of Ch10q molecules from control shRNA LOX cells displayed evidence of possible telomeric initiation (Figure S5B, molecules 1, 3–6 and 32–35), similar to the parental LOX cells, where ~29% of 10q molecules displayed evidence of possible telomeric initiation (Figure 2, molecules 1–4 and 27–31). Collectively, our SMARD results indicated that SNF2H expression leads to functional origins and replication initiation in telomeres.
Figure 6. Knockdown of SNF2H Decreases Replication Initiation within Telomeres in LOX Cells.
(A) Inducible knockdown of SNF2H in LOX cells. Immunoblot of cell lysates of LOX cell lines stably expressing Dox-inducible SNF2H shRNA or Luciferase (Luc) control shRNA, showing a time course of induction over three days.
(B) SMARD analysis of the Ch7q telomere segment from LOX cells inducibly expressing shRNA against SNF2H. Cells were grown in the presence of 1 μg/mL Dox for 48 h prior to pulse-labeling with IdU, followed by labeling with CldU in the absence of Dox. Alignments of replicated molecules fully labeled with IdU (red) and CldU (green) are shown, collected from four independent samples stretched on slides (157 fully red- and 144 fully green-labeled molecules were also collected).
(C) SMARD analysis of the Ch10q telomere segment from LOX cells inducibly expressing shRNA against SNF2H as in (B). Alignments of replicated molecules fully labeled with IdU (red) and CldU (green) are shown, collected from two independent samples stretched on slides (55 fully red- and 58 fully green-labeled molecules were also collected). Vertical lines (orange and blue) demarcate the boundaries where FISH probes bind, as described in Figure 1. Symbols are as in Figure 1. Replication profile histograms are shown under the molecule alignments.
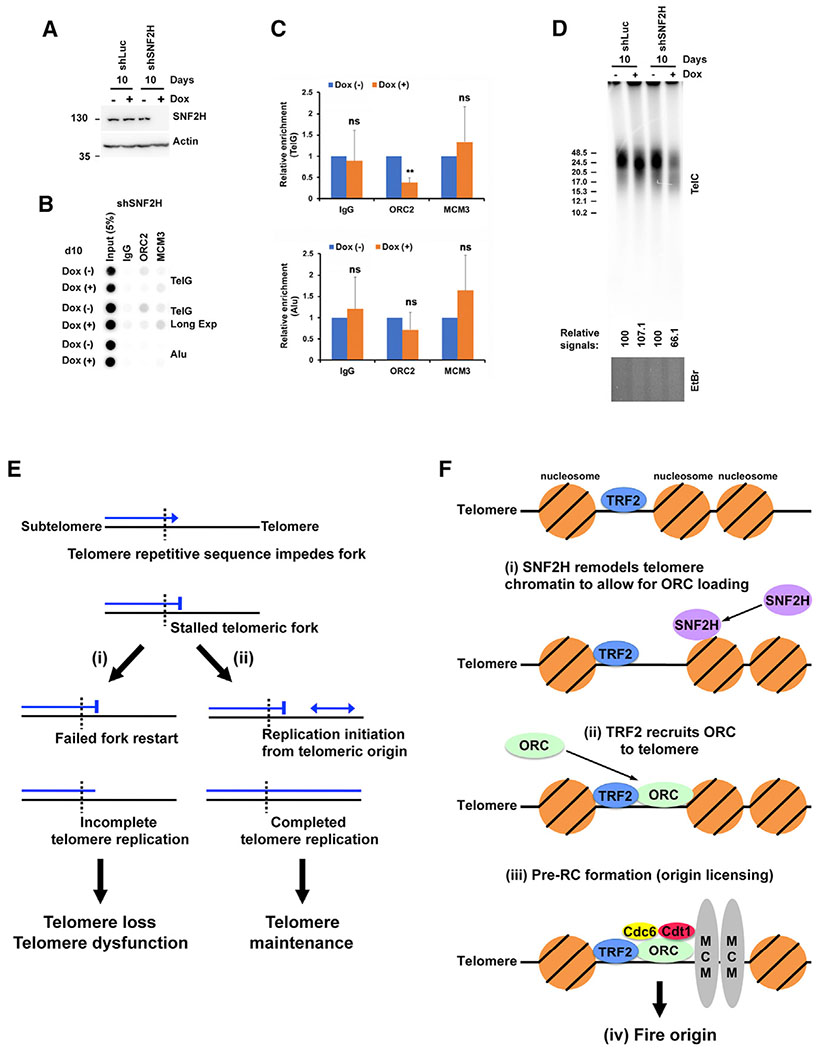
Knockdown of SNF2H Reduces ORC2 Recruitment to Telomeres and Results in Telomere Signal Loss in LOX Cells
Our SMARD results indicated that SNF2H depletion led to a significant reduction in telomeric initiation events. This suggested that SNF2H could facilitate telomeric origin formation. To explore this possibility, we examined the effect of SNF2H depletion on levels of telomere-bound ORC2 and MCM3. ChIP analysis of our LOX line expressing SNF2H-specific shRNA (see above) revealed that SNF2H depletion led to an ~63% reduction in telomere-recruited ORC2, although no change in telomere-bound MCM3 was detected (Figures 7A–7C). These data indicated that the reduction in telomeric initiation events in SNF2H depleted cells is linked to decreased ORC2 binding to telomeres. Because we found that reduction of telomeric initiation events, induced by TRF2ΔB expression was accompanied by telomere shortening and loss (Figure 4), we performed TLAs on SNF2H-depleted cells to determine whether telomere maintenance is affected in these cells. We found that the total telomere signal intensity in SNF2H depleted LOX cells was reduced by ~37% after 10 days of continuous shSNF2H expression compared to shLuc control (Figure 7D). In addition, telomere shortening was observed in the shSNF2H expressing cells in the TLAs (Figure 7D). Collectively, these data indicated that SNF2H facilitates ORC binding to telomeres and suppresses telomere repeat loss in LOX cells.
Figure 7. Knockdown of SNF2H Reduces ORC2 Recruitment to Telomeres and Results in Telomere Signal Loss in LOX Cells.
(A) Immunoblot of cell lysates of LOX cell lines stably expressing Dox-inducible SNF2H shRNA or Luc control shRNA.
(B) ChIP analysis of LOX cells stably expressing Dox-inducible SNF2H shRNA grown in the absence+ (−) or presence (+) of 1 μg/mL Dox for 10 days using antibodies against ORC2, MCM3, or IgG (control). Blots were probed by hybridization with 32P-labeled TelG or Alu probes.
(C) Quantification of four independent experiments represented in (B). The ChIP data were first normalized to input, and the percent input for Dox (+) is shown as relative to Dox (−), which was set as 1. Student’s t test was used for statistical analysis. Error bars indicate SD. **p < 0.002; ns, p > 0.05.
(D) Telomere length analysis of LOX cell lines stably expressing Dox-inducible SNF2H shRNA or Luc control shRNA. Cells were grown in the absence (−) or presence (+) of 1 μg/mL Dox for 10 days. Telomere length and relative amount of telomeric DNA were determined by restriction digestion of genomic DNA with AluI/MboI, followed by PFGE and Southern hybridization with a 32P-labeled (CCCTAA)4 probe. Ethidium bromide staining of the total DNA digest was used to normalize for DNA loading (bottom). Fragment size (in kilobases) is indicated on the left of the blot. The relative intensity of telomeric DNA signals (− Dox versus + Dox) is indicated below the blot.
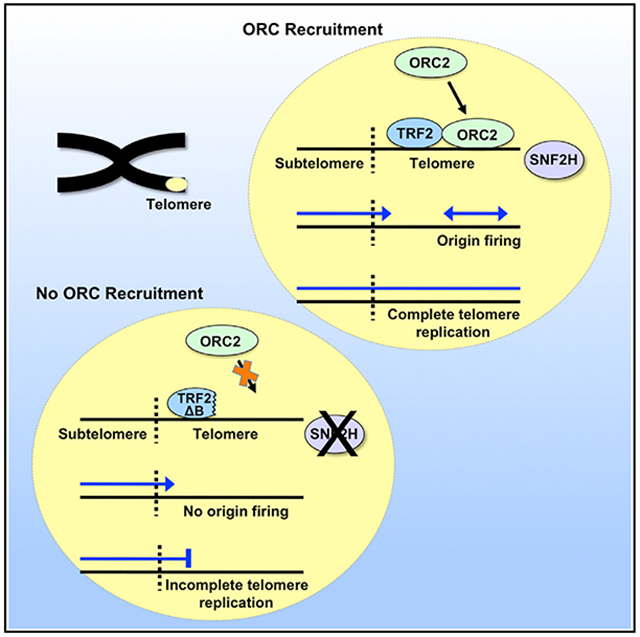
(E) Model of telomeric origin function in telomere replication. The repetitive sequence of telomeric DNA impedes and stalls replication forks. Failure to restart replication (i) leads to incomplete replication, telomere repeat loss, and dysfunction. Restart of replication from telomeric origins (ii) results in completion of telomere replication and proper telomere maintenance.
(F) Model of telomeric origin assembly and firing. (i) SNF2H remodels telomere chromatin to allow ORC loading. (ii) TRF2 recruits the ORC. (iii) The ORC recruits cdc6 and cdt1, followed by double-hexamer MCM replicative helicase loading, resulting in pre-RC formation (origin licensing). (iv) Telomeric origin fires.
DISCUSSION
TRF2 plays a direct role in telomere maintenance by forming and protecting telomeric T-loop structures (Doksani et al., 2013) that suppress DNA damage responses by preventing ataxia telangiectasia mutated (ATM) kinase signaling (Karlseder et al., 2004). TRF2 has also been shown to be indirectly involved in telomere replication via its recruitment of accessory proteins, such as RecQ helicases, that aid in removing impediments to fork movement. We have shown previously that telomeric recruitment of ORC by TRF2 facilitates formation of telomeric heterochromatin (Deng et al., 2009). Our current findings now demonstrate that TRF2 recruitment of ORC also enables telomeric initiation events, revealing a direct role of TRF2 in telomere replication. Furthermore, ORC recruitment is accompanied by telomeric recruitment of pre-RC MCM3 (Figures 3C and 3D). This is consistent with previous findings linking TRF2 to telomeric binding of pre-RC components (Bartocci et al., 2014; Tatsumi et al., 2008). Importantly, MCM3 occupancy levels at telomeres track with ORC occupancy (Figures 3C and 3D), indicating origin licensing of the telomere-bound ORC. Thus, telomeric initiation events mediated by the TRF2-recruited ORC occur at bone fide replication origins.
We showed previously that human telomere replication is most frequently accomplished by replisomes originating in the subtelomere (Drosopoulos et al., 2012). The results of these studies further indicated that telomere length appeared to be a key determinant of telomeric origin use, suggesting that infrequent use of origins within human telomeres is linked to their length. Support for this idea came from studies of mouse telomeres, which are significantly longer (>2.5×) than most human telomeres, where we identified a telomere locus (Ch14q) that is replicated primarily from origins within the telomere (Drosopoulos et al., 2015). However, our current work demonstrates that increased telomere length is not always accompanied by frequent use of telomeric origins. Although telomere replication predominantly initiates within the long Ch7q telomere of LOX cells (Figure 1), it only occasionally initiates within the long Ch10q LOX telomeres (Figure 2). Therefore, factors beyond telomere length are important for promoting telomeric origin usage. Studies have indicated that the location and efficiency of replication initiation sites are influenced by local transcription (Karnani et al., 2010; Mesner et al., 2011; Petryk et al., 2016; Valenzuela et al., 2011; Chen et al., 2019). Despite being heterochromatinized, telomeres are transcribed into telomeric repeat-containing RNA (TERRAs) from CpG island promoters located in the directly adjacent subtelomere (Azzalin et al., 2007; Deng et al., 2012; Feretzaki et al., 2019; Nergadze et al., 2009). Although Ch10q and Ch7q subtelomere TERRA promoters have similar elements, including CpG islands and CCCTC-binding factor (CTCF) binding sites, there are fundamental differences between them. The CpG islands in the Ch10q promoter are larger and have higher GC content. Consequently, the Ch10q promoter, but not the Ch7q promoter, is subject to methylation by methyltransferases, which negatively regulates Ch10q TERRA transcription (Feretzaki et al., 2019; Nergadze et al., 2009). Furthermore, TERRA transcription of Ch10q, but not Ch7q, is positively regulated by CTCF (Deng et al., 2012). Thus, differences in the local level of telomere transcription are likely and may contribute to the differences seen between Ch7q and Ch10q in telomeric origin use.
There is high risk of telomere dysfunction when telomere replication stalls and telomeres are replicated incompletely. A primary protective strategy used by cells is to avoid stalls by enlisting accessory DNA helicases to assist with telomeric fork progression when obstacles are encountered (Ahmed and Lingner, 2018; Gilson and Géli, 2007; Oganesian and Karlseder, 2009). However, when stalling occurs, mechanisms for replication recovery are essential. Along these lines, TRF2 has been shown to aid with telomeric fork recovery by assisting with relieving topological stress at forks stalled at topological barriers (Ye et al., 2010). In fact, TRF2 has been demonstrated to assist with general genomic heterochromatin replication, particularly at pericentromeric heterochromatin, by resolving topological stress at compromised forks (Mendez-Bermudez et al., 2018). In addition to these telomeric and non-telomeric roles in replication, we uncover an additional role for TRF2 in telomere replication recovery: enabling formation of telomeric origins. We have proposed previously that the ability to initiate replication within the telomere might safeguard against dysfunction by providing rescue of stalled telomere replication (Drosopoulos et al., 2012). The replication inhibition studies presented here reveal that aphidicolin-mediated slowing/stalling of forks that replicate telomere repeats induces origin firing within the repeats to complete telomere replication (Figure 5). Thus, not only are telomeric origins used as the primary origins that normally replicate certain telomeres (e.g., Ch7q; Figure 1), but they are also used to recover from compromised telomere replication (Figure 7E).
Excessively short, dysfunctional telomeres can give rise to chromosome fusion events, leading to rearrangements, deletions, or amplifications that drive genome instability. Our present work has revealed that TRF2ΔB-induced reduction of TRF2-mediated (ORC-dependent) telomeric initiation events is accompanied by telomere repeat shortening and loss and telomere dysfunction (Figure 4). Previous work has shown that TRF2ΔB expression leads to rapid telomere repeat loss because of homologous recombination (HR)-mediated T-loop deletion, resulting in formation of extrachromosomal telomeric circles (T circles) (Wang et al., 2004). However, T circle analysis by 2D gel electrophoresis showed only a slight and transient increase in T circle formation after TRF2ΔB expression in LOX cells (Figure S6), indicating that these cells are resistant to HR-mediated T loop deletion. It is possible that T circles may be unstable in LOX cells and, therefore, underdetected, although we are not aware of any studies demonstrating cell-line-specific instability of T circles. Thus, it appears that, although HR may contribute to the telomere shortening phenotype in LOX cells, the contribution is minor, particularly over longer periods of TRF2ΔB induction. This implies that telomeric origins used during normal (Figure 1) or compromised (Figure 5) telomere replication serve to suppress telomere loss and dysfunction. Interestingly, it has been observed previously that overexpression of full-length WT TRF2 leads to a decrease in telomere replication in LOX cells (Nera et al., 2015). It has been proposed that this could be the result of too much telomere-bound TRF2, creating excessive numbers of fork barriers that lead to impaired telomere replication. Importantly, although WT TRF2 overexpression is predicted to increase the number of telomeric origins, forks proceeding from telomeric “rescue” origins would still encounter and stall at TRF2 barriers. Therefore, telomeric origins may only mitigate, but not fully protect against, incomplete telomere replication resulting from excess telomere-bound WT TRF2. Unexpectedly, ectopic expression of full-length WT TRF2 showed only a slight effect on telomere replication in our studies (Figure S2). This could reflect lower levels of ectopic TRF2 expression (thus, fewer TRF2 fork barriers) in our cells compared with the previous study with LOX cells (Nera et al., 2015). Mechanistically, rapid telomere shortening could result from double-stranded DNA (dsDNA) breaks occurring at stalled telomeric forks that fail to be rescued by telomeric origins. The scarcity of origins within common fragile sites (CFS) and the resulting inability to use origins to rescue replication forks challenged by their secondary structure-forming sequences underlies the double-stranded break (DSB)-prone nature of CFSs (Durkin and Glover, 2007; Letessier et al., 2011; Li and Wu, 2020). Thus, telomeric origins appear to have a key role in protecting against telomere repeat loss and genomic instability.
Contrary to previous predictions, human telomeric heterochromatin is permissive for replication initiation (Figures 1 and 2; Drosopoulos et al., 2012). Given the compact nature of heterochromatin, it is anticipated that chromatin remodelers would be needed for opening telomeric chromatin to replication factors. The chromatin remodeler SNF2H has been found previously at human telomeres (Déjardin and Kingston, 2009) and is associated with heterochromatin replication (Collins et al., 2002). Our results indicate that SNF2H contributes to formation of functional telomeric origins (Figure 6) by providing access for ORC and possibly other initiation and/or licensing factors. Importantly, these results reveal that an active mechanism exists for establishing origins in the initiation-challenging environment of telomere chromatin. Our studies also demonstrate that SNF2H suppresses telomere repeat loss, consistent with SNF2H-facilitated telomeric initiation events providing protection against telomere loss. These findings reinforce the concept that telomeric origins protect telomeres from dysfunction and uncover a new role of SNF2H in telomere maintenance.
There are some technical limitations to our study. We focused on one cell type, LOX cells, because they have exceptionally long telomeres that are technically advantageous for SMARD analysis. In addition, we utilized a single shRNA for hSNF2H. However, this was designed using the shERWOOD algorithm (Knott et al., 2014) to reduce off-target activity.
Thus, we envision that initiation of replication within telomeres begins with recruitment of ORC proteins by TRF2. Recruitment of the ORC is facilitated by SNF2H, which remodels telomere chromatin to allow ORC loading. The telomere-bound ORC recruits Cdc6 (Borlado and Méndez, 2008; Siddiqui et al., 2013), stabilizing the ORC to chromatin, as shown in yeast (reviewed in DePamphilis, 2016). Cdt1 is recruited to the telomere-bound ORC, followed by double-hexamer MCM replicative helicase loading (origin licensing) by Cdt1 and Cdc6 (Evrin et al., 2009; Remus et al., 2009). Once licensed, the pre-RC is activated, and the origin fires to initiate replication (Figure 7F). Significantly, our data establish telomeres as the only human genomic loci demonstrated so far where active and specific assembly of origins occurs. We have shown previously that targeted recruitment of the ORC to Epstein Barr virus OriP by TRF2 can stimulate replication activity of the viral origin; however, this also requires the viral EBNA1 protein (Atanasiu et al., 2006; Deng et al., 2002). It will be of interest to determine whether there are cellular factors that substitute for EBNA1 to assist with TRF2-mediated replication initiation in the telomere.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Carl Schildkraut (schildkr@aecom.yu.edu).
Materials Availability
Transfer of materials may require a material transfer agreement (MTA) to be signed.
Data and Code Availability
This study did not generate datasets or code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture
LOX human (male) melanoma cells (Fodstad et al., 1988) were grown at 37°C and 5% CO2 in complete RPMI (high glucose RPMI supplemented with 10% FBS, 2 mM Glutamax (Fisher Scientific), 20mM HEPES, 100 I.U./mL Penicillin, and 100 μg/mL Streptomycin (Corning)). The line was authenticated by short tandem repeat (STR) profiling.
METHOD DETAILS
Plasmid construction
The doxycycline-inducible lentiviral plasmid used to express N-terminally Myc-tagged full length TRF2 or TRF2ΔB, a TRF2 mutant lacking the B domain (aa 1-44), was generated as follows. The inducible expression lentiviral vector pInducer10 (Meerbrey et al., 2011; a gift from W Guo, Albert Einstein College of Medicine, NY) was digested with AgeI and MluI, and a linker containing XhoI and KpnI-compatible BstXI sites inserted between the AgeI-MluI sites to generate pInducer10L (Drosopoulos et al., 2020). The human full length TRF2 cDNA sequence was excised from pLPC-NMYC TRF2 (gift from Titia de Lange, Addgene plasmid # 16066) and the TRF2ΔB cDNA sequence excised from pLPC-NMYC TRF2deltaB (gift from Titia de Lange, Addgene plasmid # 16067) with XhoI and KpnI and inserted into pInducer10L, directly downstream of the doxycycline-inducible promoter, to generate pIND-MYC-TRF2 and pIND-MYC-TRF2ΔB, respectively.
To generate the doxycycline-inducible SNF2H shRNA-expressing construct pIndSNF2HshmiR, an shRNA hairpin sequence targeting SNF2H, TGCTGTTGACAGT GAGCGCCCAGTTCTTTCCTCCACGTTATAGTGAAGCCACAGATGTATAACGTGGAGGAAAGAACTGGATGCCTACTGCCTCGGA, was inserted into the miR30 scaffold of the pInducer10 vector by PCR. A non-targeting control shRNA-expressing construct, pIndLucshmiR, was generated by inserting a Renilla luciferase-targeting shRNA hairpin sequence, TGCTGTTGACAGTGAGCGCAGGAATTATAATGCTTATCTATAGTGAAGCCACAGATGTATAGATAAGCATTATAATTCCTATGCCTACTGCCTCGGA, into the pInducer10 vector. These shRNAs were designed using the shERWOOD algorithm, which selects rare shRNA designs that are highly specific and highly potent at single copy representation in the genome (Knott et al., 2014).
All constructs were sequenced to confirm that unintended mutations were not introduced during PCR and cloning.
Generation of stable cell lines
Stable LOX cells inducibly expressing full-length (FL)TRF2 and TRF2ΔB protein or shRNAs were generated by lentiviral transduction. Cells were infected with lentiviral particles containing pInducer10L, pIND-MYC-TRF2, pIND-MYC-TRF2ΔB, pIndSNF2HshmiR or pIndLucshmiR, and single cell clonal colonies were selected in 1 μg/mL puromycin. N-terminally Myc-tagged FL-TRF2 and TRF2ΔB protein or shRNAs (SNF2H or Luc) were expressed in stable lines by induction with 1 μg/ml doxycycline.
Immunoblotting
Cells were harvested by trypsinization, suspended in complete RPMI, washed with PBS, then pelleted and flash frozen in liquid N2 and stored at −80°C. For SDS-PAGE, pellets were thawed on ice and lysed by resuspending in Laemmli Bufffer (60 mM Tris-HCl pH 6.8, 400 mM 2-mercaptoethanol, 2% SDS, 10% glycerol, 0.01% bromophenol blue) to a final concentration of 106 cells ml−1. Lysates were denatured (5min/100°C) and passed through a 25-gauge needle (5x) then spun for 2 min at full speed in a microfuge. Aliquots of lysate corresponding to 105 cells were resolved on 4%–15% gradient SDS-PAGE gels (BioRad), proteins transferred to nitrocellulose membrane and blocked in PBS with 5% Blotting-grade Blocker (BioRad) and 0.1% Tween-20. Membranes were then incubated with primary antibodies diluted in PBS with 5% Blotting-grade Blocker. Primary antibodies used were: anti-Myc tag (mouse monoclonal, Cell Signaling), anti-human TRF2 (rabbit polyclonal, Novus), anti-human SNF2H (mouse monoclonal, Santa Cruz), anti-α-tubulin (mouse monoclonal, Sigma), and anti-actin (rabbit polyclonal, Sigma). Following incubation with primary antibodies membranes were washed with PBS + 0.1% Tween 20. Membranes were then incubated with fluorescently-labeled Goat Anti-Mouse IRDye 680LT (Li-Cor) and Goat Anti-Rabbit IRDye800CW (Li-Cor) secondary antibodies then washed in PBS + 0.1% Tween 20. Antibody signal was detected using Luminata HRP detection reagent (Millipore). Immunoblots were then imaged on an Odyssey Lc Infrared scanner (Li-Cor) or Luminescent Imager 680 (Amersham Biosciences).
SMARD
SMARD was performed essentially as previously described (Drosopoulos et al., 2012; Figure 1). Exponentially growing cells were sequentially pulse labeled with 30 μM IdU (4 h) followed by 30 μM CldU (4 h). In experiments examining the effects of aphidicolin treatment, cells were exposed to aphidicolin (Sigma) (final [110 nM]) during the 4 h IdU labeling pulse only. Following pulsing, labeled cells were embedded in 0.5% low melting agarose (InCert, FMC) at 106 cells per 80 μl agarose cell plug and lysed overnight at 50°C in 1% n-lauroylsarcosine, 0.5 M EDTA pH 8 containing 20 mg/ml proteinase K. The plugs were then washed with TE (10 mM Tris pH 8, 1 mM EDTA), treated with 200 μM of phenylmethanesulfonyl fluoride (PMSF) then washed with TE pH 8. Subsequently, the plugs were equilibrated in restriction enzyme digestion buffer (New England Biolabs) then Pme I (30u/plug) was added and the DNA in the plugs digested in situ by overnight incubation at 37°C. The digested plugs were cast into 0.7% gels (SeaPlaque GTG, Lonza) and the DNA separated by pulse field gel electrophoresis (PFGE) using a CHEF-DRII system (Bio-Rad). Specific telomere/subtelomere chromosome segments within the gel were then located by Southern blotting (PCR list probes). Pulse field gel slices containing the telomere/subtelomere segments of interest were excised and melted and the DNA in the gel solution stretched on microscope slides coated with 3-aminopropyltriethoxysilane (Sigma). The stretched DNA was denatured in alkali buffer (0.1 M NaOH in 70% ethanol and 0.1% β-mercaptoethanol) and fixed in alkali buffer containing 0.5% glutaraldehyde. The denatured, fixed DNA was hybridized overnight with biotinylated probesat 37°C in humidified chamber. Biotinylated DNA FISH probes used to identify specific telomeric/subtelomeric segments: Ch 7q - nucleotides 158980894-159014530 (probe 1) and 159037147-159082858 (probe 2) and Ch 10q – nucleotides 135365825-135402175 (probe 1) and 135429047-135466998 (probe2)(all map coordinates based on NCBI Human Genome Map Build 37.3) were prepared by nick-translation in presence of biotin-16-dUTP (Lifetech). A biotin-OO-(CCCTAA)4 PNA probe (50 nM) (BioSythesis) was used to hybridize to the G-rich strand to identify the telomeric portion of the segments. Following hybridization, the slides were blocked with 1% BSA for a minimum of 20 min. FISH probes were then detected by incubating with an Alexa Fluor 350–conjugated NeutrAvidin (Invitrogen) followed by two rounds of incubation first with a biotinylated anti-avidin antibody (Vector) and then the Alexa Fluor 350–conjugated NeutrAvidin. The two incorporated, halogenated nucleosides were visualized by indirect immunostaining, during the second round of FISH detection, using a mouse anti-IdU monoclonal antibody (Becton Dickinson) and a rat anti-CldU monoclonal antibody (Accurate) followed by Alexa Fluor 568-conjugated goat anti-mouse (ThermoFisher) and Alexa Fluor 488-conjugated goat anti-rat (ThermoFisher) antibodies.
Photomicrograph image acquisition and analysis
Images of immunostained molecules were acquired at room temperature using a Zeiss Axioskop 2 fluorescence microscope equipped with a Plan Apochromatic 63X 1.4 NA oil objective and a charge-coupled device camera (CoolSNAP HQ; Photometrics) using IPLab software (BD). Images were processed with Photoshop (Adobe) and aligned according to the FISH probe pattern using Illustrator software (Adobe). Only images of molecules that were fully labeled with IdU, CldU or both IdU and CldU, and displayed signals of both FISH probes and telomere-specific PNA probe were collected. Replication analysis, i.e., determination of locations of origins and direction of fork movement and termination sites, by SMARD was based on analysis of the mixed red and green (IdU/CldU) dual labeled molecules. Only the dual labeled IdU/CldU molecules, which have red-green transitions, contain unambiguous information on the temporal order of replication in the DNA segment. The transitions indicate that red (IdU) labeled regions (which were labeled during the first pulse) replicated before green (CldU) labeled regions (which were labeled during the second pulse). The red and green labeling patterns reveal and map the position, direction, and density of the replication forks in the replicated molecules. Although not used for replication analysis, molecules fully labeled in only red (IdU) or only green (CldU) provide a valuable internal control. They provide a relative measure of DNA replication during each labeling pulse. Approximately equal numbers of fully red and fully green molecules obtained in a SMARD experiment indicate that replication proceeded at the same rate during both labeling pulses.
ChIP assays
ChIP assays were performed as described previously (Deng et al., 2007). Briefly, ChIP DNA at telomeres was quantitated by dot blotting with probes specific for telomere repeat DNA or Alu repeat. ChIP DNA was denatured, dot blotted onto GeneScreen Plus blotting membranes (Perkin-Elmer) and UV crosslinked at 125 mJ. Oligonucleotide probes telomere repeats ((TTAGGG)4 or (TAACCC)4) or Alu repeats (CGGAGTCTCGCTCTGTCGCCCAGGCTGGAGTGCAGTGGCGCGA) were labeled with γ-[32P] ATP (3000 Ci/mmol) and T4 nucleotide kinase (New England Biolabs). The membrane was prehybridized in Church hybridization buffer for 1 h at 42°C. A heat-denatured 32P-labeled probe was added and hybridized at 42°C overnight. Membrane was washed twice for 5 min each with 0.2 M wash buffer (0.2 M Na2HPO4 pH 7.2, 1 mM EDTA, and 2% SDS) at room temperature and once for 10 min with 0.1 M wash buffer at 42°C, developed with an Amersham Typhoon 9410 Imager (GE Healthcare) and quantified with ImageQuant TL software (GE Healthcare). Antibodies used in ChIP assays include rabbit polyclonal antibodies ORC2 (Bethyl), MCM3 (Abcam), histone H3K9me3 (Diagenode) or IgG (Cell Signaling). Rabbit antibodies to TRF2 were generated against recombinant protein and affinity purified (Deng et al., 2009). Statistical analysis of ChIP measurements was performed using Excel to calculate the mean, SDs, and p values by using a paired Student’s t test.
Telomere length analysis
Telomere length assay was performed as described previously (Deng et al., 2007) with some modifications. Briefly, genomic DNA was isolated from cells using a genomic DNA purification kit (Promega) and digested with AluI and MboI. Digested genomic DNA was purified by phenol/chloroform extraction and quantified using a NanoDrop spectrophotometer (Thermal Scientific). Equal amounts of digested DNA (5 μg) were separated by PFGE performed with a 1% agarose gel at 6V/cm, switch times from 1 to 6 s, 14°C for 13 h using CHEF-DRIII system (Bio-Rad). The resolved DNA was vacuum dried at 50°C for 90 minutes using a Model 583 Gel Dryer (Bio-Rad), in-gel denatured and neutralized and analyzed by Southern blot. The blots were hybridized with a 32P-labeled CCCTAA repeat probe at 42°C overnight, then washed twice for 5 min each with 0.2 M wash buffer (0.2 M Na2HPO4 pH 7.2, 1 mM EDTA, and 2% SDS) at room temperature and once for 10 min with 0.1 M wash buffer at 42°C. Relative telomere-repeat signals were determined by Typhoon 9410 Imager and ImageQuant TL software (GE Healthcare). Signal intensity represents the percentage decrease of average telomere signals relative to dox (−) control.
Metaphase Telomere FISH
FISH analysis on metaphase spreads was performed as previously described (Deng et al., 2007). Briefly, LOX TRF2ΔB cells cultured in the absence or presence of doxycycline (1 μg/ml) for 20 days were subcultured into fresh medium and incubated at 37°C for 24 h. Colcemid (0.1 mg/ml; GIBCO) was then added to cells and incubated for 4 h to accumulate mitotic cells. Cultures were trypsinized (Trypsin-EDTA; GIBCO), and cells were suspended in 75 mM KCl hypotonic solution at 37°C for 25 min before fixation in fresh 3:1 methanol/acetic acid for 4-5 times. Fixed cells were dropped onto cold, wet glass microscope slides and allowed to dry slowly in a humid environment. Metaphase chromosome spreads were fixed in 4% paraformaldehyde in 1x PBS for 3 min, dehydrated in ethanol series (70%, 95%, 100% (vol/vol)), and air-dried. Slides were denatured for 5 min at 81°C in hybridization mix (70% (vol/vol) formamide, 10 mM Tris·HCl (pH 7.2), and 0.5% blocking solution (Roche)) containing telomeric PNA-Cy3-(CCCTAA)3 probe (PNA Bio). After denaturation, hybridization was continued for 2 h at room temperature in the dark. Slides were washed twice for 15 min with 70% (vol/vol) formamide, 10 mM Tris·HCl (pH 7.2), and 0.1% BSA, and then three times for 5 min each with 0.15 M NaCl, 0.1 M Tris·HCl (pH 7.2), and 0.08% Tween-20. Nuclei were counterstained with 0.1 μg/mL DAPI in 1x PBS and slides were mounted with VectorShield (Vector Laboratories). Images were acquired at room temperature with a Nikon 80i Upright microscope (Nikon Instruments) equipped with a 100x lens, using ImagePro Plus software (Media Cybernetics) for image capture and processing. All images were taken at same exposure settings. Statistical analysis of telomere aberrations was performed using a two-tailed Student’s t test. Quantitative FISH analysis of metaphase spread images was performed using NIS Elements Advanced Research (ver.5.21, Nikon). Briefly, spot detection tool and Nikon AI were used to identify telomeres, and restrictions were applied to remove objects with max intensity obj > 254 or objects with touching borders from count. Count data was exported into Excel as area, number of objects, mean, and sum Intensity. Sum intensity from each metaphase FISH was used to generate histogram and graph using GraphPad Prism8 software. Statistical analysis was performed using Mann-Whitney analysis.
2D Neutral/Neutral Gel Electrophoresis
Genomic DNA was isolated using a genomic DNA isolation kit (Promega) and digested with AluI and MboI. Two-dimensional gel electrophoresis was performed as previously described (Deng et al., 2013). Equal amounts of digested DNA (10–15 μg) were subjected to electrophoresis in a 0.4% agarose gel (first dimension) at 30V for 12–14 h at room temperature and then in a 1.2% agarose gel (second dimension) containing 0.3 mg/ml ethidium bromide at 150V for 6 h at 4°C. The gel was then transferred to a GeneScreen Plus blotting membrane (Perkin-Elmer) and analyzed by Southern blotting as described for telomere length analysis assay. Telomere DNA was visualized by overnight hybridization with a 32P-labeled (TTAGGG)4 probe at 42°C. Telomeric signals were quantified using ImageQuant TL software (GE Healthcare). Telomeric circles (T circle) ratio was calculated by taking the ratio of the circular telomeric DNA signal to the linear telomeric DNA signal as previously described (Deng et al., 2007).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
Statistical analysis of ChIP measurements and telomere aberrations were performed using Excel (Microsoft, Redmond, WA) to calculate the mean, SDs, and p values. P values were determined using a two-tailed Student’s t test. Statistical analysis of metaphase spread sum intensities was performed using Prism 8 software (GraphPad, San Diego, CA) to calculate the mean, SDs, and p values. P values of pairwise comparisons were determined using a Mann-Whitney test. Statistical details of experiments can be found in the figure legends.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Myc tag | Cell Signaling | Cat# 2276S; RRID:AB_331783 |
| Rabbit polyclonal anti-human TRF2 | Novus | Cat# NB110-57130; RRID:AB_844199 |
| Mouse monoclonal anti-human SNF2H | Santa Cruz | Cat# sc-365727; RRID:AB_10844618 |
| Goat biotinylated anti-avidin | Vector | Cat#BA-0300; RRID:AB_2336108 |
| Mouse monoclonal anti-BrdU(IdU) | BD | Cat# 347580; RRID:AB_400326 |
| Rat monoclonal anti-BrdU(CldU) | Accurate | Cat# OBT0030; RRID:AB_2313756 |
| Rabbit polyclonal anti-actin | Sigma | Cat#A2066; RRID:AB_476693 |
| Goat polyclonal anti-Mouse IgG Alexa Fluor 568 | ThermoFisher | Cat#A11031; RRID:AB_144696 |
| Goat polyclonal anti-Rat IgG Alexa Fluor 488 | ThermoFisher | Cat#A11006; RRID:AB_2534074 |
| Goat polyclonal anti-Rabbit IRDye800CW | Li-Cor | Cat#926-32211; RRID:AB_621843 |
| Goat polyclonal anti-Mouse IRDye 680LT | Li-Cor | Cat#926-68020; RRID:AB_10706161 |
| Rabbit polyclonal anti-ORC2 | Bethyl | Cat# A302-735A: RRID:AB_10627808 |
| Rabbit polyclonal anti-MCM3 | Abcam | Cat#ab4460; RRID:AB_304469 |
| Rabbit polyclonal anti-H3K9me3 | Diagenode | Cat#C15410193; RRID:AB_2616044 |
| Rabbit polyclonal anti-TRF2 | Deng et al., 2009 | N/A |
| Normal rabbit IgG | Cell Signaling | Cat#2729S; RRID:AB_1031062 |
| Bacterial and Virus Strains | ||
| Escherichia coli Stable competent cells | NEB | Cat#C3040H |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Doxycycline hyclate | Sigma | Cat#D9891 |
| Puromycin | Sigma | Cat#P8833 |
| Aphidicolin | Sigma | Cat# A0781 |
| Q5 Hot Start High-Fidelity DNA Polymerase | NEB | Cat#M0493 |
| XtremeGene HP transfection reagent | Roche | Cat#06 366 244 001 |
| Alexa Fluor 350–conjugated NeutrAvidin | Invitrogen | Cat# A11236 |
| Colcemid | GIBCO | Cat# |
| Blocking Reagent | Roche | Cat#11096176001 |
| Blotting-grade Blocker | BioRad | Cat# 1706404 |
| Critical Commercial Assays | ||
| QIAquick PCR Purification Kit | QIAGEN | Cat#28104 |
| QIAquick Gel Extraction Kit | QIAGEN | Cat#28704 |
| Wizard Genomic DNA Purification Kit | Promega | Cat# A1120 |
| Experimental Models: Cell Lines | ||
| LOX human melanoma cells | Laboratory of Lifeng Xu | RRID:CVCL_1381 |
| Oligonucleotides | ||
| Primer: shRNA Forward: ACGTGCGGCCGCTCCAGCCCTCACTCCTTCTC | This paper | N/A |
| Primer: shRNA Reverse: CAAAGAACGGAGCCGGTTGG | This paper | N/A |
| Biotin-OO-(CCCTAA)4 PNA | BioSythesis | N/A |
| Cy3-(CCCTAA)3 PNA | PNA Bio | N/A |
| Recombinant DNA | ||
| pLPC-NMYC TRF2 | Smogorzewska and de Lange, 2002 | Addgene Plasmid # 16066; RRID:Addgene_16066 |
| pLPC-NMYC TRF2deltaB | Smogorzewska and de Lange, 2002 | Addgene Plasmid #16067; RRID:Addgene_16067 |
| pInducer10 | Meerbrey et al., 2011 | Addgene Plasmid #44011; RRID:Addgene_44011 |
| pInducer10L | Drosopoulos et al., 2020 | N/A |
| pIND-MYC-TRF2 | This paper | N/A |
| pIND-MYC-TRF2ΔB | This paper | N/A |
| pIndSNF2HshmiR | This paper | N/A |
| pIndLucshmiR | This paper | N/A |
| Software and Algorithms | ||
| ImageStudio | Li-Cor | https://www.licor.com/bio/image-studio/ |
| ImageJ | NIH | https://imagej.nih.gov |
| ImageQuant TL | GE Healthcare | http://www.cytivalifesciences.com/country-selection?originalItemPath=%2f |
| ImagePro Plus | Media Cybernetics | https://www.mediacy.com/imageproplus |
| NIS Elements Advanced Research | Nikon | https://www.microscope.healthcare.nikon.com/products/software/nis-elements |
| Excel | Microsoft | https://www.microsoft.com/en-us/microsoft-365?rtc=1 |
| Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Photoshop | Adobe | https://www.adobe.com/products/photoshop.html |
| Illustrator | Adobe | https://www.adobe.com/products/illustrator.html |
Highlights.
Decreased TRF2-mediated ORC2 recruitment reduces replication initiation in telomeres
Telomeric origins are activated by replication stress to complete telomere replication
Reduction of telomere initiation events leads to telomere repeat loss and dysfunction
Depletion of SNF2H decreases ORC2 recruitment and replication initiation in telomeres
ACKNOWLEDGMENTS
We thank Ken Chang (Cold Spring Harbor Laboratories Cancer Center Functional Genomics & Genetics core) for SNF2H- and Renilla luciferase-targeting shRNA sequences. We also thank Frederick Keeney (Wistar Imaging core facility) for image quantification and Samantha Soldan for assistance with GraphPad software. This work was supported by National Institutes of Health/National Institute of General Medical Sciences grant 5R01-GM045751 (to C.L.S.) and National Institutes of Health/National Cancer Institute grant 5R01-CA140652 (to P.M.L.). S.T. was supported by training grant T-32 NIH 5T32AG023475.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108379.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Ahmed W, and Lingner J (2018). Impact of oxidative stress on telomere biology. Differentiation 99, 21–27. [DOI] [PubMed] [Google Scholar]
- Anand RP, Shah KA, Niu H, Sung P, Mirkin SM, and Freudenreich CH (2012). Overcoming natural replication barriers: differential helicase requirements. Nucleic Acids Res. 40, 1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasiu C, Deng Z, Wiedmer A, Norseen J, and Lieberman PM (2006). ORC binding to TRF2 stimulates OriP replication. EMBO Rep. 7, 716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, and Lingner J (2007). Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318, 798–801. [DOI] [PubMed] [Google Scholar]
- Bartocci C, Diedrich JK, Ouzounov I, Li J, Piunti A, Pasini D, Yates JR 3rd, and Lazzerini Denchi E (2014). Isolation of chromatin from dysfunctional telomeres reveals an important role for Ring1b in NHEJ-mediated chromosome fusions. Cell Rep. 7, 1320–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH, Epel ES, and Lin J (2015). Human telomere biology: A contributory and interactive factor in aging, disease risks, and protection. Science 350,1193–1198. [DOI] [PubMed] [Google Scholar]
- Blasco MA (2007). The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet 8, 299–309. [DOI] [PubMed] [Google Scholar]
- Borlado LR, and Méndez J (2008). CDC6: from DNA replication to cell cycle checkpoints and oncogenesis. Carcinogenesis 29, 237–243. [DOI] [PubMed] [Google Scholar]
- Chen YH, Keegan S, Kahli M, Tonzi P, Fenyö D, Huang TT, and Smith DJ (2019). Transcription shapes DNA replication initiation and termination in human cells. Nat. Struct. Mol. Biol 26, 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins N, Poot RA, Kukimoto I, García-Jiménez C, Dellaire G, and Varga-Weisz PD (2002). An ACF1-ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin. Nat. Genet 32, 627–632. [DOI] [PubMed] [Google Scholar]
- Déjardin J, and Kingston RE (2009). Purification of proteins associated with specific genomic Loci. Cell 136, 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Lezina L, Chen CJ, Shtivelband S, So W, and Lieberman PM (2002). Telomeric proteins regulate episomal maintenance of Epstein-Barr virus origin of plasmid replication. Mol. Cell 9, 493–503. [DOI] [PubMed] [Google Scholar]
- Deng Z, Dheekollu J, Broccoli D, Dutta A, and Lieberman PM (2007). The origin recognition complex localizes to telomere repeats and prevents telomere-circle formation. Curr. Biol 17, 1989–1995. [DOI] [PubMed] [Google Scholar]
- Deng Z, Norseen J, Wiedmer A, Riethman H, and Lieberman PM (2009). TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell 35, 403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Wang Z, Stong N, Plasschaert R, Moczan A, Chen HS, Hu S, Wikramasinghe P, Davuluri RV, Bartolomei MS, et al. (2012). A role for CTCF and cohesin in subtelomere chromatin organization, TERRA transcription, and telomere end protection. EMBO J. 31, 4165–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Glousker G, Molczan A, Fox AJ, Lamm N, Dheekollu J, Weizman O-E, Schertzer M, Wang Z, Vladimirova O, et al. (2013). Inherited mutations in the helicase RTEL1 cause telomere dysfunction and Hoyeraal-Hreidarsson syndrome. Proc. Natl. Acad. Sci. USA 110, E3408–E3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis ML (2016). Genome Duplication: The Heartbeat of Developing Organisms. Curr. Top. Dev. Biol 116, 201–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doksani Y, Wu JY, de Lange T, and Zhuang X (2013). Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell 155, 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosopoulos WC, Kosiyatrakul ST, Yan Z, Calderano SG, and Schildkraut CL (2012). Human telomeres replicate using chromosome-specific, rather than universal, replication programs. J. Cell Biol 197, 253–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosopoulos WC, Kosiyatrakul ST, and Schildkraut CL (2015). BLM helicase facilitates telomere replication during leading strand synthesis of telomeres. J. Cell Biol 210, 191–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosopoulos WC, Vierra DA, Kenworthy CA, Coleman RA, and Schildkraut CL (2020). Dynamic Assembly and Disassembly of the Human DNA Polymerase 5 Holoenzyme on the Genome In Vivo. Cell Rep. 30, 1329–1341.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin SG, and Glover TW (2007). Chromosome fragile sites. Annu. Rev. Genet 41, 169–192. [DOI] [PubMed] [Google Scholar]
- Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, and Speck C (2009).Adouble-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNAreplication. Proc. Natl. Acad. Sci. USA 106, 20240–20245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feretzaki M, Renck Nunes P, and Lingner J (2019). Expression and differential regulation of human TERRA at several chromosome ends. RNA 25, 1470–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodstad O, Aamdal S, McMenamin M, Nesland JM, and Pihl A (1988). A new experimental metastasis model in athymic nude mice, the human malignant melanoma LOX. Int. J. Cancer 41, 442–449. [DOI] [PubMed] [Google Scholar]
- Ge XQ, Jackson DA, and Blow JJ (2007). Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 21, 3331–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E, and Géli V (2007). How telomeres are replicated. Nat. Rev. Mol. Cell Biol. 8, 825–838. [DOI] [PubMed] [Google Scholar]
- Higa M, Kushiyama T, Kurashige S, Kohmon D, Enokitani K, Iwahori S, Sugimoto N, Yoshida K, and Fujita M (2017). TRF2 recruits ORC through TRFH domain dimerization. Biochim. Biophys. Acta Mol. Cell Res 1864, 191–201. [DOI] [PubMed] [Google Scholar]
- Hyrien O (2015). Peaks cloaked in the mist: the landscape of mammalian replication origins. J. Cell Biol 208, 147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa AS, Zhou JQ, Schulz VP, Monson EK, and Zakian VA (2002). Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 16, 1383–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlseder J, Hoke K, Mirzoeva OK, Bakkenist C, Kastan MB, Petrini JH, and de Lange T (2004). The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol. 2, E240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani N, Taylor CM, Malhotra A, and Dutta A (2010). Genomic study of replication initiation in human chromosomes reveals the influence of transcription regulation and chromatin structure on origin selection. Mol. Biol. Cell 21, 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott SRV, Maceli A, Erard N, Chang K, Marran K, Zhou X, Gordon A, Demerdash OE, Wagenblast E, Kim S, et al. (2014). A computational algorithm to predict shRNA potency. Mol. Cell 56, 796–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letessier A, Millot GA, Koundrioukoff S, Lachagès AM, Vogt N, Hansen RS, Malfoy B, Brison O, and Debatisse M (2011). Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature 470, 120–123. [DOI] [PubMed] [Google Scholar]
- Levenson V, and Hamlin JL (1993). A general protocol for evaluating the specific effects of DNA replication inhibitors. Nucleic Acids Res. 21, 3997–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, and Wu X (2020). Common fragile sites: protection and repair. Cell Biosci 10, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalingam D, Tay LL, Tan WH, Chai JH, and Wang X (2011). Mutant telomerase RNAs induce DNA damage and apoptosis via the TRF2-ATM pathway in telomerase-overexpressing primary fibroblasts. FEBS J 278, 3724–3738. [DOI] [PubMed] [Google Scholar]
- Makovets S, Herskowitz I, and Blackburn EH (2004). Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol. Cell. Biol 24, 4019–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez P, and Blasco MA (2017). Telomere-driven diseases and telomere-targeting therapies. J. Cell Biol 216, 875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerbrey KL, Hu G, Kessler JD, Roarty K, Li MZ, Fang JE, Herschkowitz JI, Burrows AE, Ciccia A, Sun T, et al. (2011). The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc. Natl. Acad. Sci. USA 108, 3665–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Bermudez A, Lototska L, Bauwens S, Giraud-Panis MJ, Croce O, Jamet K, Irizar A, Mowinckel M, Koundrioukoff S, Nottet N, et al. (2018). Genome-wide Control of Heterochromatin Replication by the Telomere Capping Protein TRF2. Mol. Cell 70, 449–461.e5. [DOI] [PubMed] [Google Scholar]
- Mesner LD, Valsakumar V, Karnani N, Dutta A, Hamlin JL, and Bekiranov S (2011). Bubble-chip analysis of human origin distributions demonstrates on a genomic scale significant clustering into zones and significant association with transcription. Genome Res. 21, 377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Rog O, and Cooper JP (2006). Semi-conservative DNA replication through telomeres requires Taz1. Nature 440, 824–828. [DOI] [PubMed] [Google Scholar]
- Miotto B, Ji Z, and Struhl K (2016). Selectivity of ORC binding sites and the relation to replication timing, fragile sites, and deletions in cancers. Proc. Natl. Acad. Sci. USA 113, E4810–E4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, and Cech TR (1997). Telomerase catalytic subunit homologs from fission yeast and human. Science 277, 955–959. [DOI] [PubMed] [Google Scholar]
- Nera B, Huang HS, Lai T, and Xu L (2015). Elevated levels of TRF2 induce telomeric ultrafine anaphase bridges and rapid telomere deletions. Nat. Commun 6, 10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nergadze SG, Farnung BO, Wischnewski H, Khoriauli L, Vitelli V, Chawla R, Giulotto E, and Azzalin CM (2009). CpG-island promoters drive transcription of human telomeres. RNA 15, 2186–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norio P, and Schildkraut CL (2001). Visualization of DNA replication on individual Epstein-Barr virus episomes. Science 294, 2361–2364. [DOI] [PubMed] [Google Scholar]
- Oganesian L, and Karlseder J (2009). Telomeric armor: the layers of end protection. J. Cell Sci 122, 4013–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryk N, Kahli M, d’Aubenton-Carafa Y, Jaszczyszyn Y, Shen Y, Silvain M, Thermes C, Chen CL, and Hyrien O (2016). Replication landscape of the human genome. Nat. Commun 7, 10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, and Diffley JF (2009). Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell 139, 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samassekou O, Gadji M, Drouin R, and Yan J (2010). Sizing the ends: normal length of human telomeres. Ann. Anat 192, 284–291. [DOI] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, and de Lange T (2009). Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138, 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui K, On KF, and Diffley JF (2013). Regulating DNA replication in eukarya. Cold Spring Harb. Perspect. Biol 5, a012930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, and de Lange T (2002). Different telomere damage signaling pathways in human and mouse cells. EMBO J. 21, 4338–4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumi Y, Ezura K, Yoshida K, Yugawa T, Narisawa-Saito M, Kiyono T, Ohta S, Obuse C, and Fujita M (2008). Involvement of human ORC and TRF2 in pre-replication complex assembly at telomeres. Genes Cells 13, 1045–1059. [DOI] [PubMed] [Google Scholar]
- Urban JM, Foulk MS, Casella C, and Gerbi SA (2015). The hunt for origins of DNA replication in multicellular eukaryotes. F1000Prime Rep. 7, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela MS, Chen Y, Davis S, Yang F, Walker RL, Bilke S, Lueders J, Martin MM, Aladjem MI, Massion PP, and Meltzer PS (2011). Preferential localization of human origins of DNA replication at the 5′-ends of expressed genes and at evolutionarily conserved DNA sequences. PLoS ONE 6, e17308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdun RE, and Karlseder J (2006). The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell 127, 709–720. [DOI] [PubMed] [Google Scholar]
- Wang RC, Smogorzewska A, and de Lange T (2004). Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119, 355–368. [DOI] [PubMed] [Google Scholar]
- Woodward AM, Göhler T, Luciani MG, Oehlmann M, Ge X, Gartner A, Jackson DA, and Blow JJ (2006). Excess Mcm2-7 license dormant origins of replication that can be used under conditions of replicative stress. J. Cell Biol 173, 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Lenain C, Bauwens S, Rizzo A, Saint-Léger A, Poulet A, Benarroch D, Magdinier F, Morere J, Amiard S, et al. (2010). TRF2 and apollo cooperate with topoisomerase 2alpha to protect human telomeres from replicative damage. Cell 142, 230–242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate datasets or code.