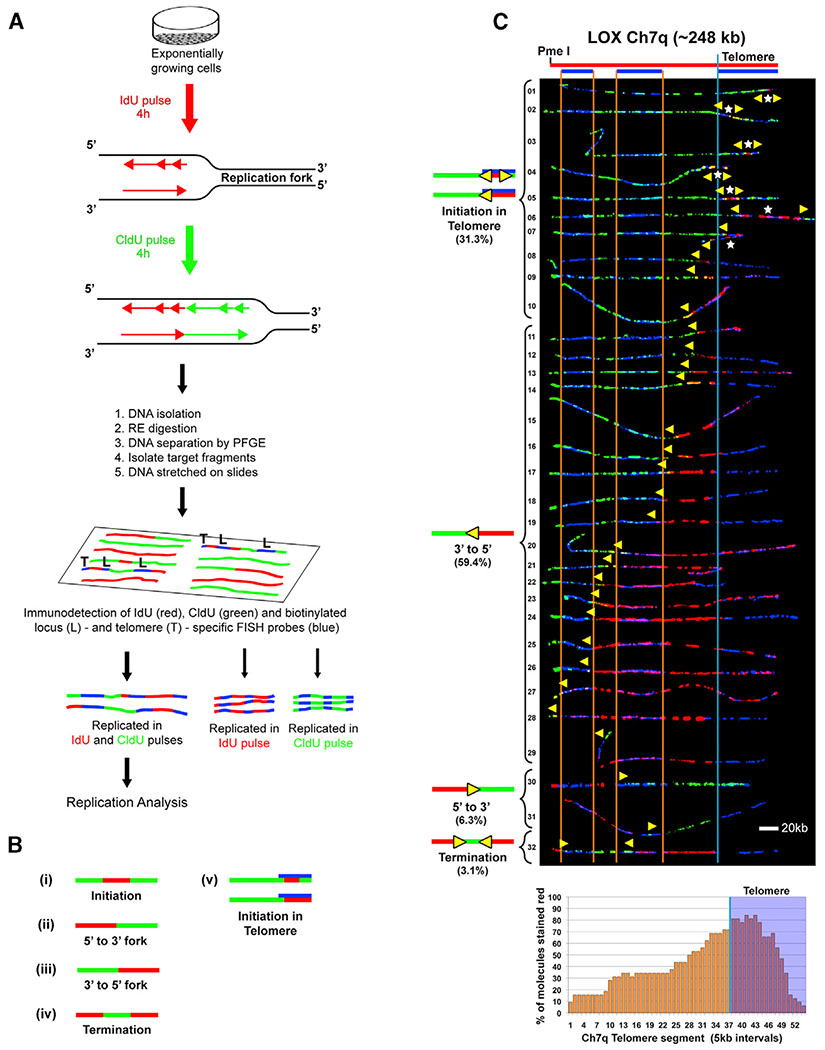
Figure 1. Telomere Replication Predominantly Initiates within the Ch7q Telomere in Human LOX Cells.
(A) SMARD. Replicating DNA in exponentially and asynchronously growing cells is labeled sequentially by pulsing with IdU for 4 h, followed by CldU for 4 h. Labeled cells are embedded in agarose, lysed, and deproteinized to release genomic DNA. The genomic DNA is digested with a rare-cutting restriction endonuclease to obtain unique large molecules, which are separated by pulsed-field gel electrophoresis (PFGE) to enrich for by size for the particular segments of interest. PCR screening is performed to identify the target fragment within the gel, which is excised. The gel slice containing the segment of interest is melted, and the enriched DNA in the melted gel solution is stretched on silanized glass slides. Indirect immunofluorescence with antibodies against the halogenated nucleosides is used to identify regions where IdU (red) or CldU (green) was incorporated into the DNA. Immunodetected biotinylated probes (blue) are used to identify the molecules to be analyzed (Drosopoulos et al., 2012). Images of molecules fully labeled with IdU, CldU, or both IdU and CldU and that also display all probes are collected. Molecules fully labeled with red and green (IdU and CldU) are used for replication analysis, whereas fully red (IdU) or fully green (CldU) molecules serve as labeling controls.
(B) Staining patterns of molecules indicate the direction of replication fork progression because replication in IdU occurs first. Molecules where one side is stained red and the other side is stained green indicate that forks progressed in a single direction through the segment (ii and iii). Initiation events are observed as a red tract flanked on both sides by green (i), whereas a green tract flanked on both sides by red indicates a termination event (iv). In addition to a red tract flanked on both sides by green, initiation events in the telomere (v) can appear as a red tract that does not extend out of the telomere or extends only a few kilobases out from the telomere.
(C) SMARD analysis of a segment of Ch7q containing the telomere from LOX cells. Alignments of replicated molecules fully labeled with IdU (red) and CldU (green) are shown, collected from four independent samples stretched on slides (156 fully red- and 164 fully green-labeled molecules were also collected). A map of the 7q locus is depicted above the alignment, with the positions of the FISH probes (blue bars below) used to identify and orient the molecules indicated. Vertical orange lines mark positions of the ends of the subtelomeric FISH signals used to align the molecules. The boundary between the subtelomere and telomere is delineated by a vertical blue line. Yellow arrows mark sites of transition from IdU to CldU incorporation and indicate the direction of fork progression at the moment of transition during replication (polarity of fork direction assigned relative to the polarity of the telomere G-rich strand of the segment). Initiation events (red tracts flanked by green) are seen in telomeres. White stars indicate the centers of initiations. A replication profile, a histogram of the percentages of molecules containing red staining (IdU) per 5-kb interval along the segment, is shown under the alignment. The 5-kb intervals that have the highest percentage of molecules stained red are those that, on average, replicate first. The telomere portion of the segment is shaded in blue, and the telomere-subtelomere boundary is indicated by a blue vertical line.