Figure 2. tRNA Fragments Accumulate in Some Mutant Strains.
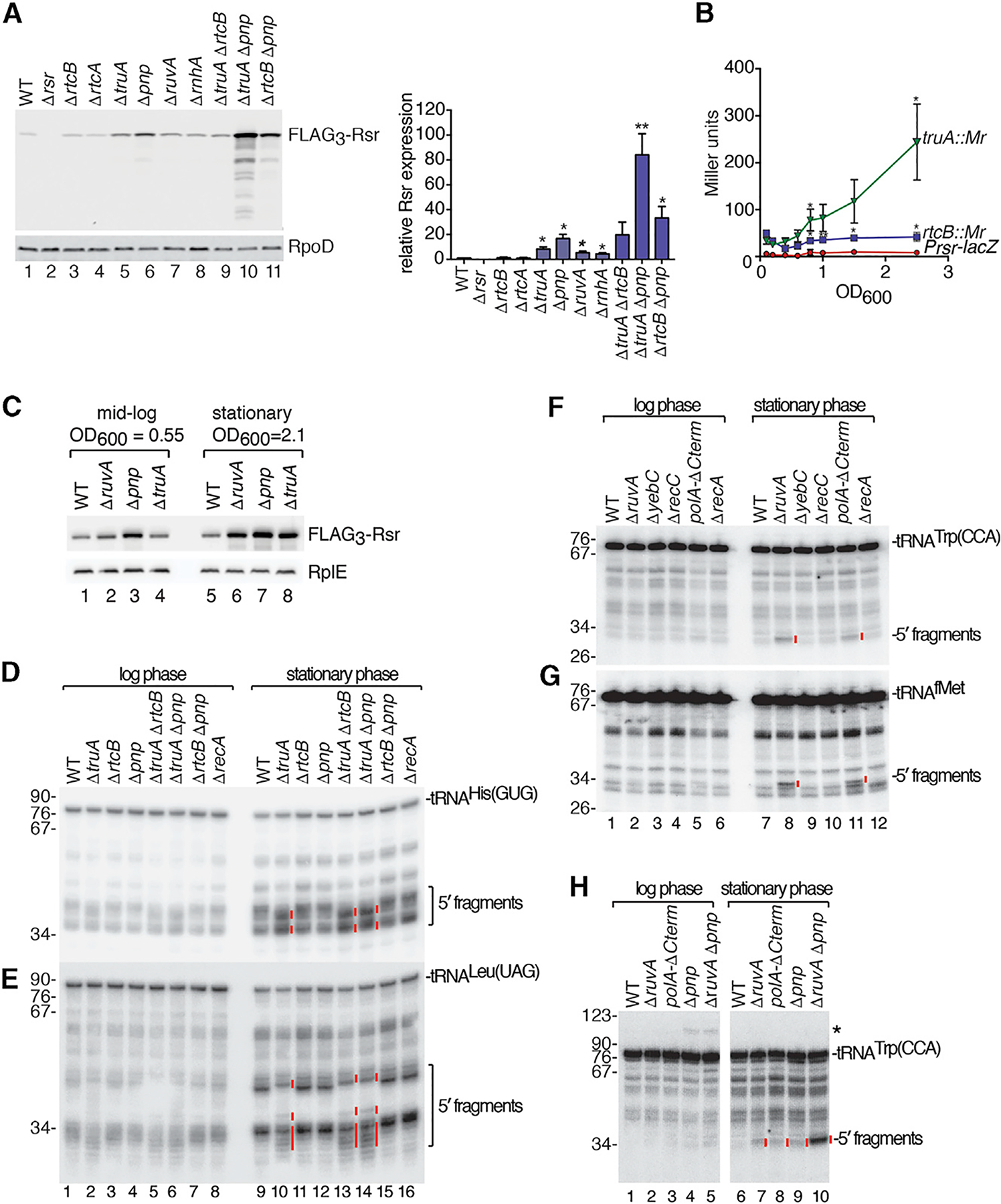
(A) 14028s FLAG3-rsr strains carrying the indicated deletions were subjected to immunoblotting to detect FLAG3-Rsr. RpoD, loading control. Right, quantitation (n = 3). p values are relative to the wild-type strain.
(B) β-Galactosidase activity was assayed in the indicated Prsr-lacZ strains as a function of growth. p values are relative to the Prsr-lacZ strain.
(C) Lysates of the indicated FLAG3-rsr strains were subjected to immunoblotting to detect FLAG3-Rsr. RplE, loading control.
(D and E) RNA from the indicated strains was subjected to northern blotting to detect 5′ halves of tRNAHis(GUG) (D) and tRNALeu(UAG) (E).
(F and G) RNA from the indicated strains was subjected to northern blotting to detect 5′ halves of tRNATrp(CCA) (F) and tRNAfMet (G).
(H) RNA from the indicated strains was probed to detect 5′ halves of tRNATrp(CCA). Asterisk denotes a tRNA precursor.
Data in (A) and (B) are mean (n = 3) ± SEM. p values were calculated with two-tailed unpaired t tests. *p < 0.05 and **p < 0.01.
In (D)–(H), red lines denote fragments unique to mutant strains in stationary phase. See also Figure S2.
