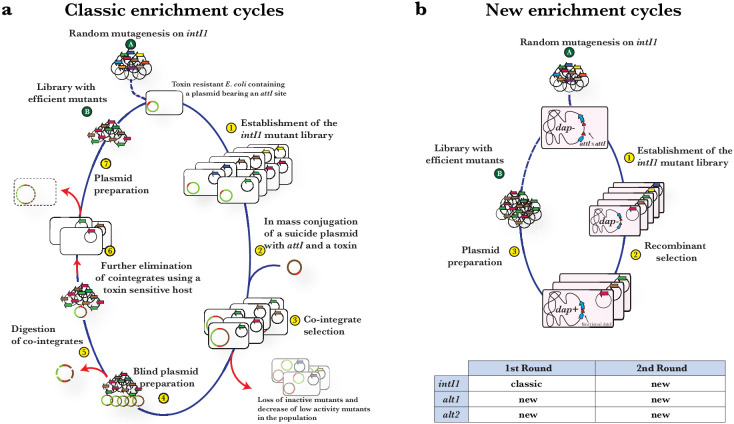
Appendix 1—figure 1. Diagram of the enrichment cycles used to select for integrase variants hyperactive for attI x attI.
(a) Cycles adapted from Demarre et al., 2007 used for the evolution experiments with intI1. Briefly, a randomized library of integrase encoding genes cloned in a pBAD plasmid (A) is established in a toxin-resistant E. coli strain containing a pSU plasmid that encodes an attI site and a toxin (1). This strain acts as receptor in the conjugation of an attI-bearing suicide plasmid (2). Mutagenized integrases deliver the attI x attI recombination reaction allowing the formation of a selectable cointegrate (3). pBAD plasmids are then purified in two steps: the plasmid preparation from recombinants (4) is digested to specifically degrade cointegrates (5) and then transformed in a toxin sensitive strain (6). Plasmid extraction from this strain yields very pure pBAD plasmids containing a variety of integrase-coding genes of high activity (B). Higher selective pressure is applied by subjecting these plasmids to further cycles. b: Novel enrichment cycles used for the evolution of the rest of alleles. Briefly: the library of integrase mutants cloned in a pBAD plasmid (A) is established in a dap- E. coli that contains a dapA gene in the chromosome interrupted by two attI sites (1). Expression of the integrase leads to the recombination of both sites and the reconstitution of dapA, allowing recombinants to grow in media not supplemented with DAP (2). Plasmid preparations from recombinants (3) yields pure pBAD plasmids containing a mixture of hyperactive-integrase-coding genes (B) that can be further used in subsequent cycles.