Extended Data Fig. 6. H3K27me3 and MLL2 dependent transcription.
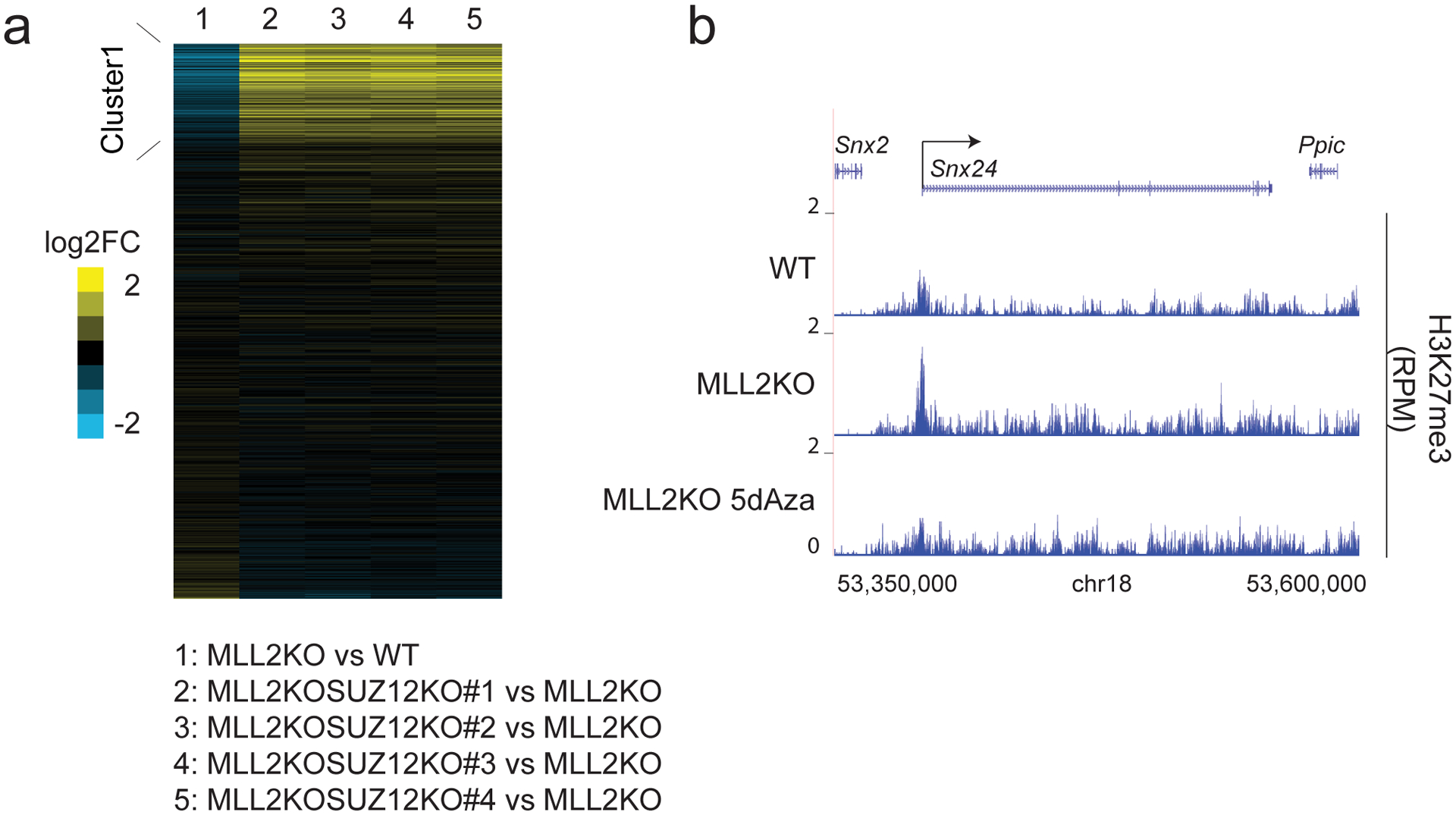
(a) Heatmaps show the corresponding log2 fold changes in gene expression in 1) MLL2KO vs. WT mESCs, 2) MLL2KOSUZ12KO#1 vs. MLL2KO mESCs, 3) MLL2KOSUZ12KO#2 vs. MLL2KO mESCs, 4) MLL2KOSUZ12KO#3 vs. MLL2KO mESCs and 5) MLL2KOSUZ12KO#4 vs. MLL2KO mESCs. The experiment was repeated two times independently with similar results. (b) ChIP-seq tracks of H3K27me3 occupancy at the Snx24 locus in WT, MLL2KO and MLL2KO 100 nM 5dAza for 4 days treated mESCs. The experiment was repeated two times independently with similar results.
