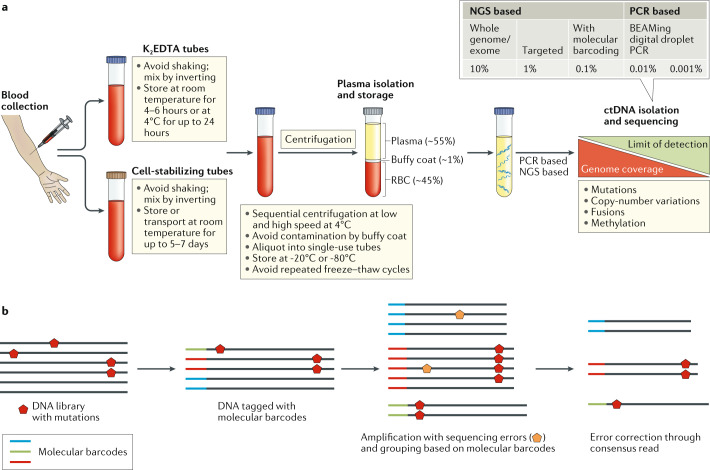
Fig. 2. ctDNA isolation and analysis.
a | Circulating cell-free DNA, of which circulating tumour DNA (ctDNA) is a part of, is isolated from plasma samples after serial centrifugation of blood collected in either K2EDTA or cell-stabilizing tubes. ctDNA is subsequently isolated from cell-free DNA using library preparations and analysed for the presence of various genetic aberrations, including mutations, copy-number variations, fusions and/or other alterations such as changes in DNA methylation. b | Molecular barcoding: prior to PCR and sequencing, each DNA fragment can be labelled with unique DNA barcodes; subsequently, reads that share the same barcode (typically in thousands) can be grouped together because they all originate from the same ctDNA fragment. This approach enables sequencing errors (orange pentagon, seen in the minority) to be distinguished from true mutations (red pentagon, seen in the majority). Molecular barcoding also helps correct potential biases in amplification and thus assists in the precise quantification of mutations or amplification frequencies. NGS, next-generation sequencing; RBC, red blood cells.