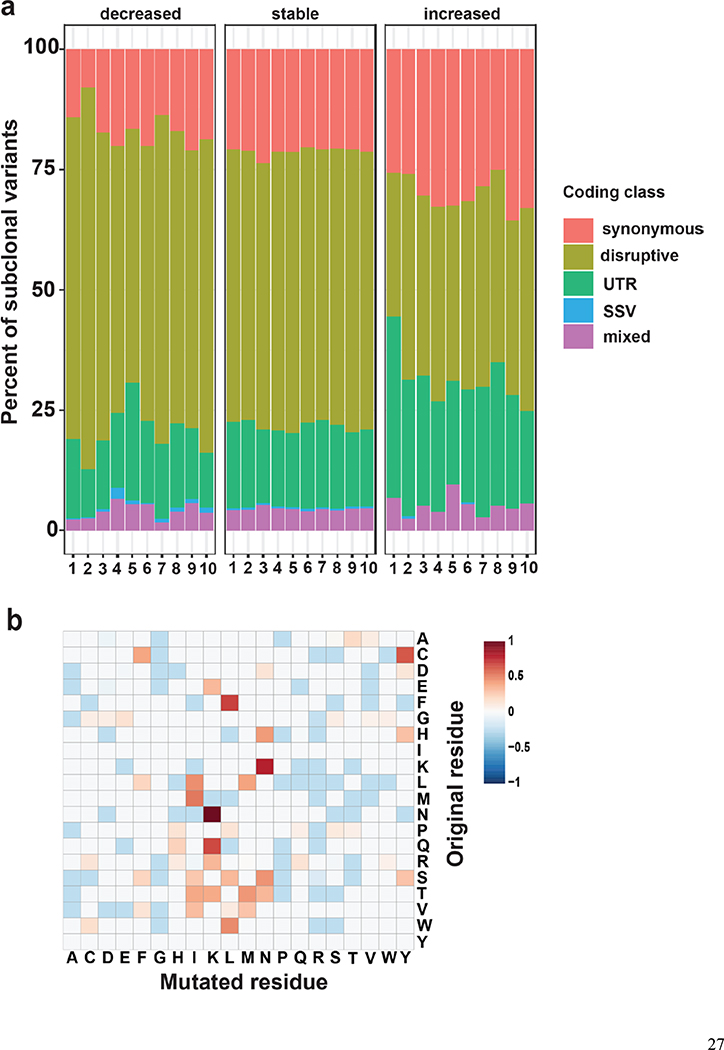
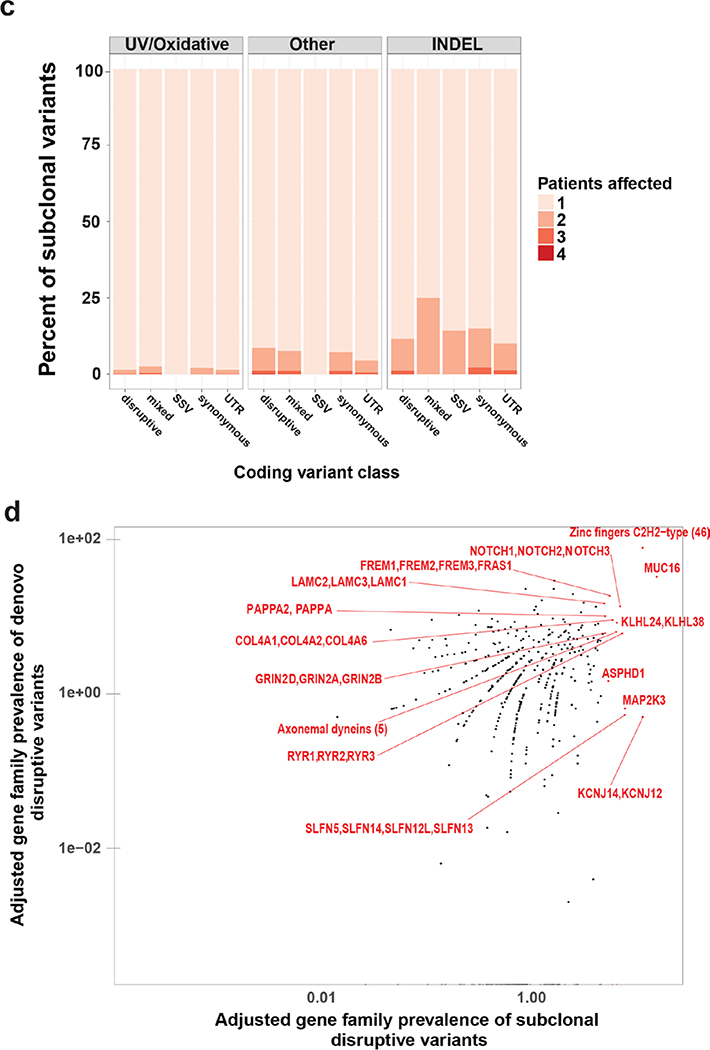
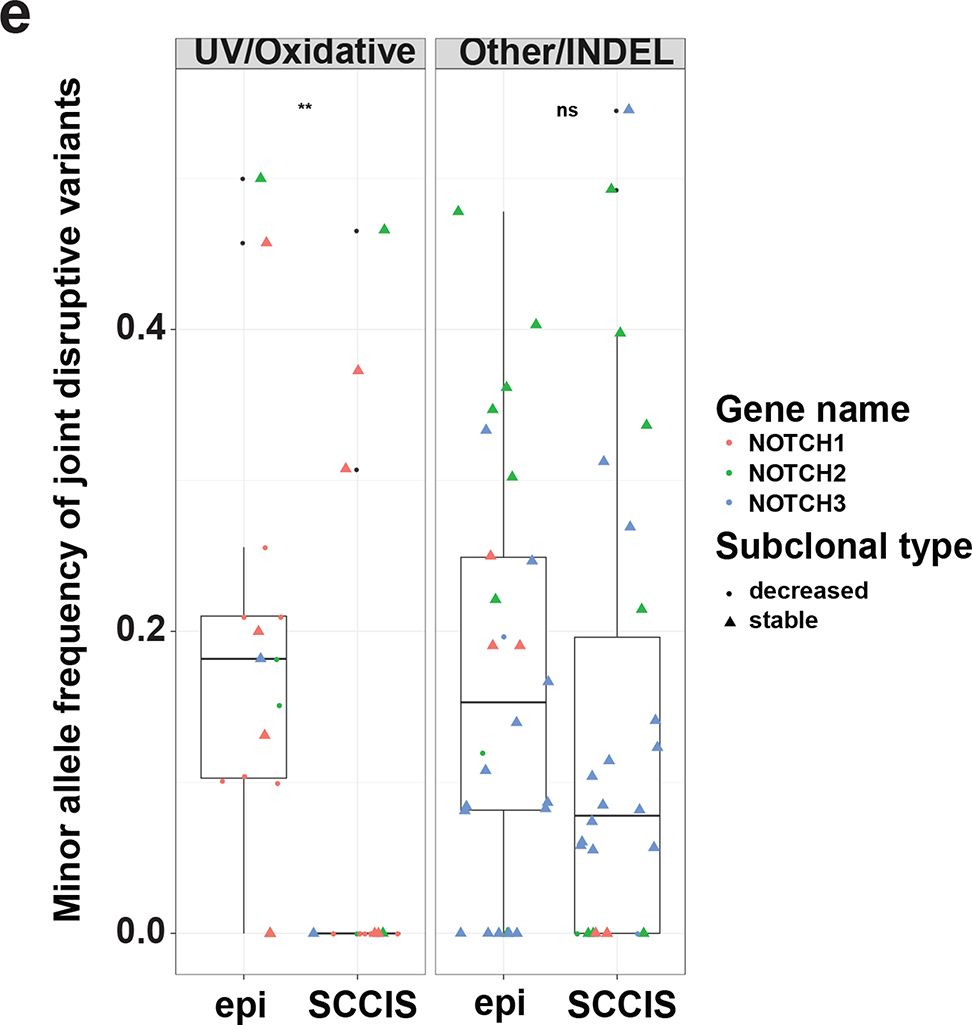
Figure 3.
Characteristics of subclonal variants. (a) Summary of the coding class for subclonal and “stable” variants. disruptive: nonsynonymous, nonsense, readthrough or frame-shift types; SSV: splice-site variant; mixed: classified into more than one coding classes; stable: similar to subclonal variants but the MAF is not significantly different between epidermis and SCCIS; (b) Relative preference of amino acid substitution preference of nonsynonymous subclonal variants using nonsynonymous stable variants as background. Rows and columns represent the original and mutated amino acids, respectively. Values shown in cells are calculated LOD scores (in bits). LOD scores for non-observed substitutions were set to 0. (c) Commonality of subclonal variants reflected as number of shared patients regarding different coding class and variant types. UV: C->T(G->A) signature variant; oxidative: G->T(C->A) signature variant; INDEL: insertion/deletions; (d) Adjusted gene-family preference of subclonal (x-axis) and de-novo (y-axis) variants. Red highlighted dots: top 15 most preferred gene families for subclonal variants with all affected genes labeled. Large families with many affected genes are labeled with family names and number of affected genes in parentheses instead. (e) Distribution of the MAF of all subclonal and stable variants for different variant signatures (UV/oxidative, Other, INDEL) using the NOTCH gene family as an example. ** indicates statistically a significant difference with p-value < 0.01 based on Wilcoxon Rank-Sum Test, ns-indicates a non-significant difference with p-value > 0.05.