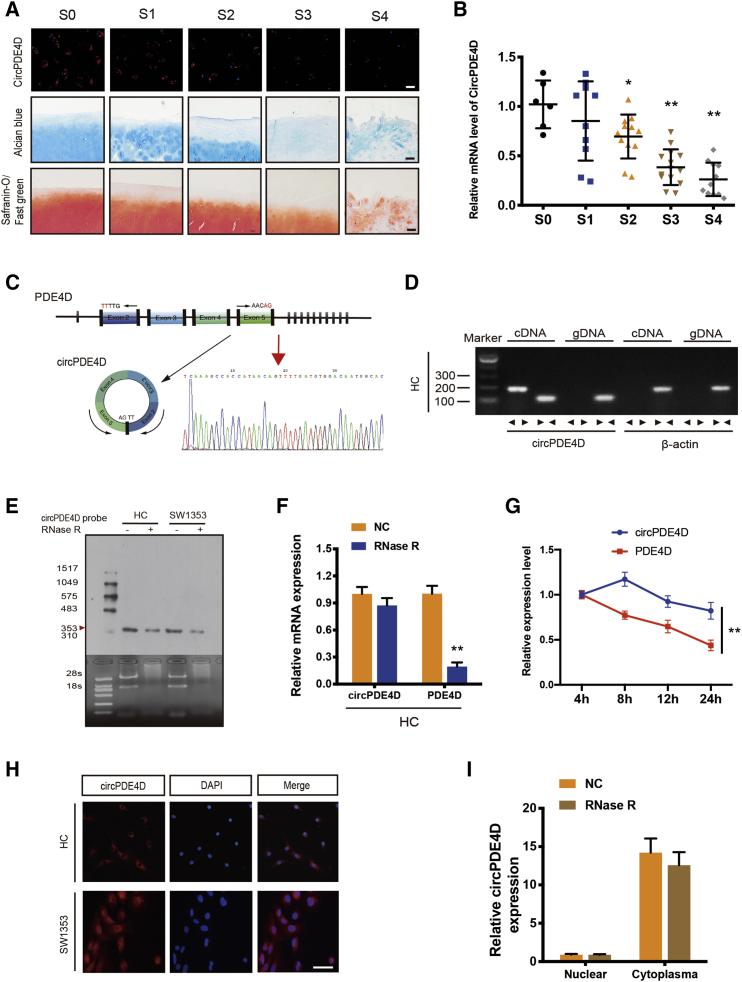
Figure 1.
circPDE4D Validation and Expression in OA Cartilage Tissue and Chondrocytes
(A) Representative images of RNA fluorescence in situ hybridization (FISH), Safranin-O/Fast green staining and Alcian blue staining in human cartilage tissues from stage 0 (S0) to stage 4 (S4). Scale bar, 50 μm. (B) The expression of circPDE4D in human cartilage specimens from stage 0 (S0) to stage 4 (S4) (n = 53) was evaluated by qRT-PCR. (C) Schematic illustration demonstrating the circularization of PDE4D exon 2–5 to form circPDE4D (black arrow). The presence of circPDE4D was validated by RT-PCR followed by Sanger sequencing. The red arrow represents “head-to-tail” circPDE4D splicing sites. (D) The presence of circPDE4D in human HCs was validated by RT-PCR. Divergent primers amplified circPDE4D from cDNA but not from genomic DNA; β-actin served as the negative control. (E) Northern blots for detecting circPDE4D in HCs and SW1353 cells treated with or without RNase R. The upper panels show the probed blots of circPDE4D, and the red triangle represents the circPDE4D band size (353 bp). The lower panels show the gel electrophoresis results obtained for RNA with or without RNase R digestion. (F) The expression of circPDE4D and linear PDE4D mRNA in HCs treated with or without RNase R was detected by qRT-PCR. The relative levels of circPDE4D and PDE4D mRNA were normalized to the value obtained with the mock treatment. (G) The levels of circPDE4D and PDE4D in HCs treated with actinomycin D at the indicated time points were detected by qRT-PCR. (H) FISH showed that circPDE4D was predominantly localized in the cytoplasm. The circPDE4D probes were labeled with CY3, and nuclei were stained with DAPI. Scale bar, 20 μm. (I) circPDE4D was detected in different cell fractions. Nuclear and cytoplasmic RNA was extracted, and junction primers were used for circPDE4D detection. U6 was used as an internal control for nuclear RNA, and β-actin was used as an internal control for cytoplasmic RNA. The data were obtained from three independent experiments with three independent donors (presented as the means ± SDs; F, G, and I) or were representative of three independent experiments with similar results (A, B, D, E, and H). ∗p < 0.05 and ∗∗p < 0.01 versus the control or indicated group. The data were analyzed by one-way ANOVA followed by the Bonferroni test (B) and two-tailed t tests (F, G, and I).