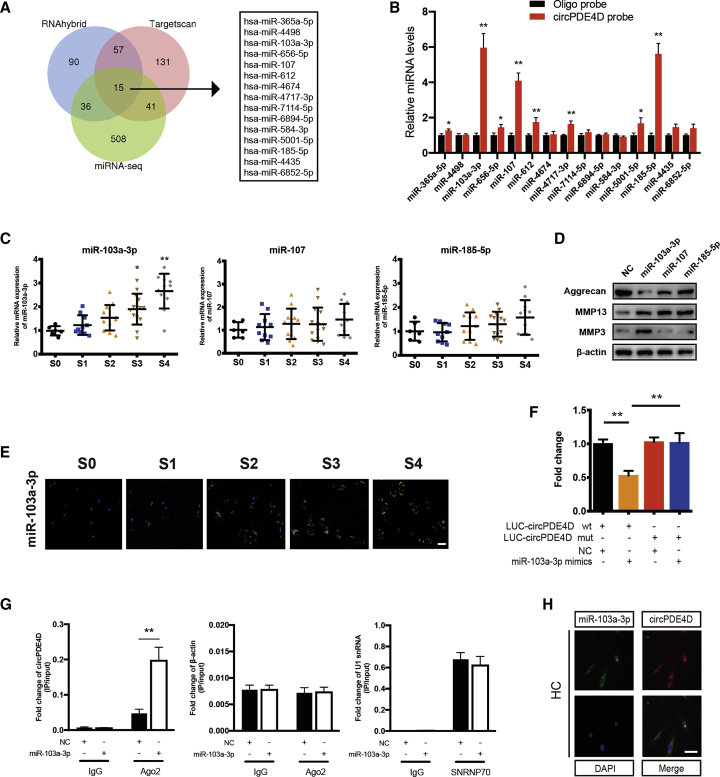
Figure 3.
circPDE4D Serves as a Sponge for miR-103a-3p in Human Chondrocytes
(A) Schematic illustration showing the overlap of the potential target miRNAs of circPDE4D predicted using the RNAhybrid and TargetScan databases and the miRNA sequences. (B) The relative levels of 15 miRNA candidates in HC lysates after pull-down assay were examined by qRT-PCR. Multiple miRNAs were pulled down by circPDE4D. (C) The expression of miR-103a-3p, miR-107, and miR-185-5p in human cartilage tissues (stage 0 to stage 4) was evaluated by qRT-PCR (n = 53). (D) HCs were transfected with miR-103a-3p, miR-107, miR-185a-5p, or negative control and then used for detection of Aggrecan, MMP3, and MMP13 proteins. (E) Representative images of FISH demonstrating the relative expression of miR-103a-3p in human cartilage tissues (stages 0–4). The corresponding Safranin-O/Fast green staining and Alcian blue staining are shown in Figure 1A. Scale bar, 50 μm. (F) HEK293T cells were transfected with miR-103a-3p (or NC) and circPDE4D reporter (or circPDE4D mut reporter) and then used for luciferase activity detection. (G) The circPDE4D levels in SW1353 cells transfected with NC or miR-103a-3p were detected by Ago2 RNA immunoprecipitation assay (RIP assay). IgG was used as a control, and β-actin was used as a negative control. SNRNP70 RIP for U1 snRNA was used as a positive control. (H) FISH images showing the colocalization of circPDE4D and miR-103a-3p in HCs. miR-103a-3p probes were labeled with Alexa Fluor 488. Locked nucleic acid circPDE4D probes were labeled with CY3, and nuclei were stained with DAPI. Scale bar, 20 μm. The data were obtained from three independent replicates (presented as the means ± SDs; B and G) or were obtained from three independent replicates (F) or were representative of three independent experiments with similar results (C, E, and H). ∗p < 0.05 and ∗∗p < 0.01 versus the control or indicated group. The data were analyzed by one-way ANOVA followed by the Bonferroni test (C) and two-tailed t tests (B, F, and G).