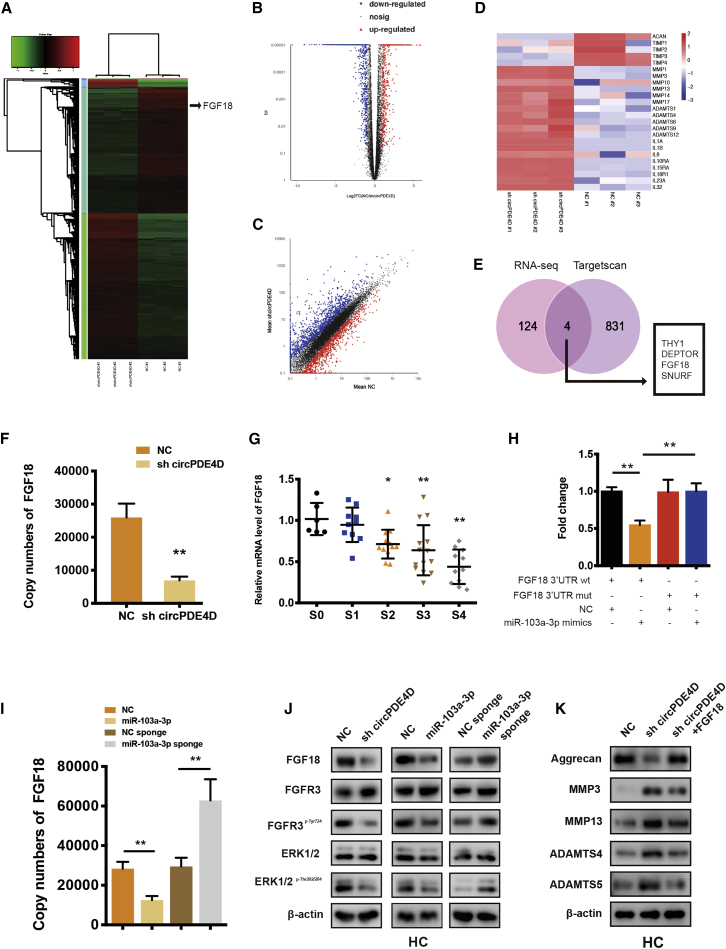
Figure 6.
FGF18 Is the Direct Target of miR-103a-3p and Mediates the Function of miR-103a-3p and circPDE4D
(A) Clustered heatmap of differentially expressed mRNAs in HCs stably transfected with NC or sh-circPDE4D. (B) In the volcano plot, significantly upregulated genes are represented by red dots, whereas significantly downregulated genes are represented by blue dots. (C) The scatter diagram shows the differently expressed genes. (D) Representative genes involved in cartilage anabolism, catabolism, or inflammation identified from the RNA-seq data. (E) Schematic diagram exhibiting the overlap between the downregulated mRNAs identified from the RNA-seq data (Log2(sh-circPDE4D/NC) < −2.5, p < 0.01) and target gene analyses of miR-103a-3p using the TargetScan database. (F) mRNA expression of FGF18 in HCs stably transfected with sh-circPDE4D. (G) The expression of FGF18 in human cartilage tissues (stage 0 to stage 4) was determined by qRT-PCR. The corresponding Safranin-O/Fast green staining and Alcian blue staining are shown in Figure 1A (n = 53). (H) HEK293T cells were cotransfected with miR-103a-3p (or NC) and luciferase reporter constructs containing wild-type (WT) or mutated FGF18 3′-UTR. The relative luciferase activity is demonstrated in the histogram. (I) Effect of miR-103a-3p on the mRNA expression of FGF18. Chondrocytes were transfected with miR-103a-3p/miR-103a-3p sponge or the corresponding NC and then used for qRT-PCR analysis. (J) The protein expression of FGF18, FGFR3, FGFR3p-Tyr724, ERK1/2, and ERK1/2p-Thr202/204 in HCs was detected by western blotting. Cells were transfected with sh-circPDE4D, miR-103a-3p, miR-103a-3p sponge, or the corresponding NC. (K) HCs stably transfected with sh-circPDE4D (or NC) or cotransfected with sh-circPDE4D and FGF18 were subjected for the western blotting detection of Aggrecan, MMP3, MMP13, ADAMTS4, and ADAMTS5. The data were obtained from three independent experiments with three independent donors (presented as the means ± SDs; F and I), were obtained from three independent replicates (H) or were representative of three independent experiments with similar results (G, J, and K). ∗p < 0.05 and ∗∗p < 0.01 versus the control or indicated group. The data were analyzed by one-way ANOVA followed by the Bonferroni test (G) and two tailed t tests (F, H, and I).