Figure 1.
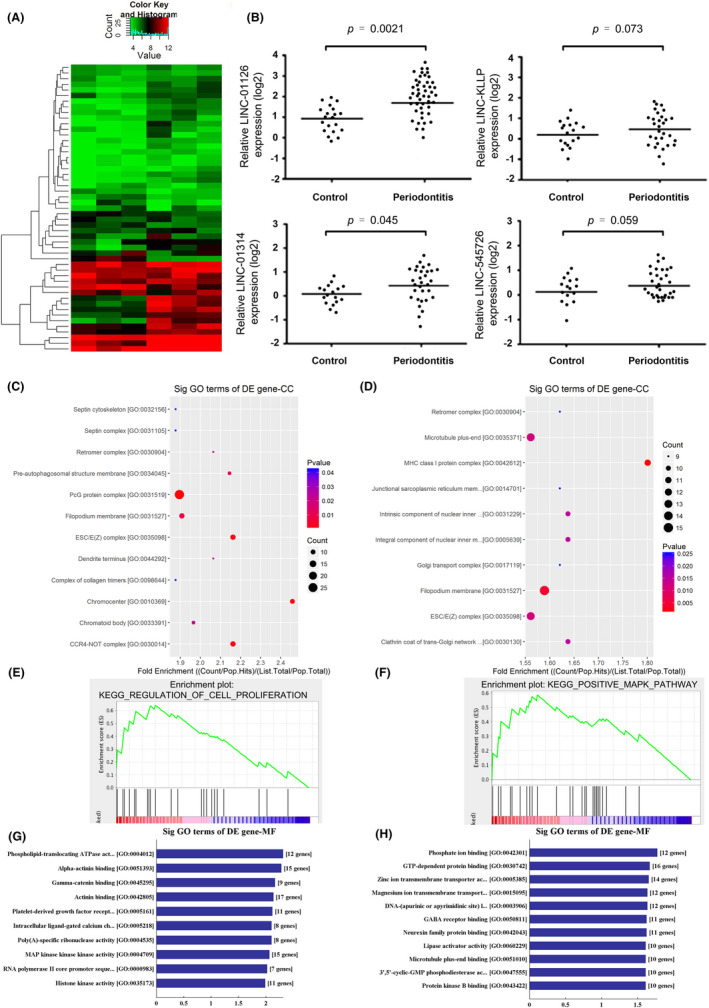
Aberrant expression of lncRNAs in periodontitis. (A) Heatmap of the top 50 differentially expressed lncRNAs (|log2FC| >1.5, P < .05) in 3 pairs of gingival tissues from patients with periodontitis and the healthy controls though lncRNA expression microarray. (B) RNA expressions of the 4 mostly upregulated lncRNAs LINC01126, LINC545726, lncRNA‐KLLP, LINC01314 in periodontal tissues and the controls group (C, D) The top 10 upregulated and downregulated GO functions of the cellular component (CC) domains. (E) GSEA plot of differentially expressed involved in proliferation in periodontal tissues. (F) GSEA plot of genes involved in MAPK signalling pathway in periodontal tissues. Running enrichment score was positive for most differentially expressed genes. (G, H) The top 10 upregulated and downregulated GO functions of the molecular function (MF) domains
