Abstract
Background:
N6-methyladenosine (m6A) is the most common form of mRNA modification under the field of “RNA epigenetics.” However, its role in ovarian cancer (OC) development is poorly understood. In the current study, we aimed to identify gene signatures and prognostic values of m6A RNA methylation regulators.
Method:
Specifically, we downloaded Mutations and Copy number variant (CNV) data from the TCGA database for 579 OC patients, then analyzed gene expression and prognosis value using integrative bioinformatics. Thereafter, we verified the related biological processes of Wilms’ tumor 1-associating protein (WTAP) gene using Gene set enrichment analysis (GSEA).
Results:
Results showed that almost all ovarian cancer patients (99.31%) have CNVs with at least 1 m6A regulatory gene, whereas 83.76% of cases exhibited concurrence of CNVs in more than 4 m6A regulatory genes. Additionally, alteration of m6A regulators was associated with historical grade, whereas integrative bioinformatics and Cox multivariate model analysis revealed a significant correlation between high WTAP expression and worse ovarian cancer outcomes. Moreover, GSEA revealed that high WTAP expression was associated with cell cycle regulation and MYC targets.
Conclusion:
Overall, our findings demonstrate the significance of high-frequency genetic alterations of m6A RNA methylation regulators and WTAP’s poor prognosis value in OC. These findings provide valuable insights into the role of m6A methylation in OC, and will be vital in guiding development of novel treatment therapies.
Keywords: bioinformatics, ovarian cancer, biomarker, N6-methyladenosine, PROGNOSIS
Introduction
Ovarian cancer (OC) is the leading cause of cancer-related deaths among all gynecological tumors.1 A large number of OC patients are diagnosed at an advanced stage, owing to lack of biomarkers for early clinical screening, as well as occurrence of relatively non-specific symptoms. Despite advances in modern management therapies, such as cytoreductive surgery and adjuvant chemotherapy, the 5-year overall survival for more than 70% of OC patients remains less than 30%.2 Therefore, there is an urgent need to identify novel factors that regulate tumorigenesis and new biomarkers for early diagnosis or prognosis.
Dysregulated gene expression is one of the hallmarks of cancer. N6-methyladenosine (m6A), one of the most dominant drivers of messenger RNA (mRNA) modification, provides a novel layer of post-transcriptional gene regulation.3,4 Previous studies have demonstrated its importance in a variety of crucial biological functions, including embryogenesis, proliferation, and differentiation of stem cells, DNA damage response, integrity of the nervous system, and adaptive stress responses.5-9 Generally, m6A regulators comprise 3 classes of components, including methylases (“writers”), demethylases (“erasers”), and m6A-binding proteins (“readers”),3,10 each playing different function. For example, the methylases complex is composed of Wilms’ tumor 1-associating protein (WTAP), methyltransferase-like 3 (METTL3), and methyltransferase-like 14 (METTL14), with METTL3 reported to be the key methyltransferase responsible for m6A modification. Obesity-associated protein (FTO) and AlkB homolog 5 (ALKBH5) belong to erasers, whereas YTH domain proteins (YTHDF1-3) and YTH domain-containing proteins (YTHDC1-2) constitute the readers. Recently, m6A regulators were implicated in formation and progression of specific malignant tumors, including hepatic malignant neoplasm,11-13 glioblastoma,14,15 acute myeloid leukemia (AML),16 breast cancer,17,18 lung cancer,19 and gastric cancer.20 However, the roles of m6A methylation in tumorigenesis and prognosis of OC remain unclear. Based on these studies, we sought to explore the gene signatures and prognostic values associated with m6A regulators in OC.
Materials and Methods
Ethics Statement
The clinicopathological information, copy number variants, mutations, gene expression microarray documents, and prognostic data were downloaded from The Cancer Genome Atlas (TCGA) project using the cBioportal website,21 Oncomine database22 and KM plotter database.23 All of these are open-access public databases. In addition, all enrolled participants provided written informed consent.
TCGA Database Analysis
We enrolled a total of 579 OC patients, with Mutations and Copy number variant (CNV) data, from the Ovarian Serous Cystadenocarcinoma (TCGA, Firehose Legacy) cohort dataset (http://www.cbioportal.org).21 Focus was given to 9 m6A regulatory genes in cbioportal, including WTAP, METTL3, METTL14, FTO, ALKBH1, ALKBH5, YTHDF1, YTHDF2 and YTHDF3. Moreover, we included the TP53 gene, which plays a critical role in the progress of OC and has an extremely high mutation rate in OC, to serve as a reference. To explore the clinical pathological and molecular parameters of diverse CNV patterns, we divided the OC cases into 2 subgroups: “With low coexisting numbers of m6A related CNV genes (≤4)” and “with high coexisting numbers of m6A related CNV genes (>4)”. We extracted and normalized RNA-Seq data using RSEM (RNA-Seq by Expectation-Maximization).
Kaplan-Meier Plotter Analysis
We then employed the Kaplan-Meier plotter online database (http://kmplot.com/analysis/),23 basing on the Gene Expression Omnibus (GEO), the European Genotype Archive (EGA) and TCGA databases for analysis of the relationship between m6A regulatory genes and prognosis in OC. During the analysis, we calculated the hazard ratio (HR), 95% confidence intervals (95%CI), and p-value.
Oncomine Database Analysis
To explore the expression patterns of the WTAP gene in OC, we employed the online tumor microarray database, Oncomine (https://www.oncomine.org/resource/login.html).22 Specifically, this database was searched using the keyword: “WTAP,” cancer type: “ovarian cancer,” and analysis type: “cancer vs. normal analysis.” Thereafter, we performed statistical comparisons using a 2-tailed Student’s t-test, with data followed by P < 0.05 regarded statistically significant.
Gene Set Enrichment Analysis (GSEA) of RNA-Seq Data
We stratified OC cases into 4 quartiles, according to patterns of WTAP expression, from the highest (fourth quartile) to the lowest (first quartile). Then, we analyzed the enriched gene sets between the lowest and highest quartiles of WTAP expression using GSEA v3.0,24 alongside MsigDB gene set (h.all.v6.0.symbols.gmt) as reference.25 To determine significant enrichment, we assumed a false discovery rate (FDR) of less than 0.25 and a p-value less than 0.05.
Statistical Analyses
Statistical analyses were performed using SPSS 19.0 (SPSS Inc, Chicago, IL, USA) and GraphPad Prism 7.0 (GraphPad Software Inc, San Diego, CA, USA) software. Specifically, correlation between the clinical-pathological parameters and diverse CNV patterns of m6A regulatory genes was analyzed using the chi-square test, and Kaplan-Meier curves generated to estimate prognosis. Data followed by P < 0.05 was regarded statistically significant.
Results
Mutations and Copy Number Variants of m6A Regulatory Genes in OC
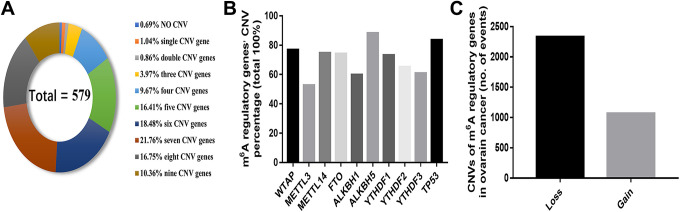
To elucidate the roles of 10 candidate m6A regulatory genes in OC development, we first analyzed genetic alterations of these genes using the cBioPortal TCGA dataset. Interestingly, we detected mutation events in m6A-related genes in 5 cases (TCGA-13-0920-01, TCGA-04-1342-01, TCGA-24-2271-01, TCGA-10-0938-01, TCGA-23-1117-01) among the 316 OC patients with mutation data (Supplementary Information 1). Notably, the mutation rate in the TP53 gene was relatively high in this cohort (87.66%, 277/316), which was consistent with a previous study.26 Additionally, among all subjects with CNV data, almost all OC patients recorded CNVs with at least 1 m6A regulatory gene (575/579, 99.31%), whereas 83.76% of the cases resulted in concurrence of CNVs in more than 4 m6A regulatory genes (Figure 1A). Furthermore, the highest frequency of CNV events was recorded in the m6A “eraser” gene ALKBH5 (88.26%, 511/579), followed by the m6A “writer” gene WTAP (76.86%, 445/579) (Figure 1B and Table 1). Furthermore, analysis of CNV patterns in OC patients revealed a higher loss (2331/3398) than gain (1067/3398) of copy number variations (Figure 1C and Table 1), which was similar to the CNV status recorded in AML and kidney malignancy.27,28
Figure 1.
CNVs of m6A regulatory genes in ovarian cancer. (A) Concurrence of CNVs in specific number m6A regulatory genes in ovarian cancer samples. (B) Frequency of ovarian cancer samples with CNVs for m6A For Peer Review regulators based on data from TCGA. (C) Events of copy number gain or loss of m6A regulatory genes in ovarian cancer samples. CNV, copy number variant. m6A, N6-methyladenosine.
Table 1.
Patterns of CNV Occurrence in 579 Ovarian Cancer Patients.
| Diploid | Deep deletion | Shallow deletion | Copy number gain | Amplification | CNV sum | Percentage | ||
|---|---|---|---|---|---|---|---|---|
| Erasers | FTO | 149 | 6 | 378 | 44 | 2 | 430 | 74.27% |
| ALKBH1 | 233 | 0 | 245 | 88 | 13 | 346 | 59.76% | |
| ALKBH5 | 68 | 8 | 484 | 19 | 0 | 511 | 88.26% | |
| Writers | METTL3 | 274 | 2 | 166 | 114 | 23 | 305 | 52.68% |
| METTL14 | 146 | 6 | 387 | 32 | 8 | 433 | 74.78% | |
| WTAP | 134 | 14 | 359 | 67 | 5 | 445 | 76.86% | |
| Readers | YTHDF1 | 155 | 0 | 28 | 315 | 81 | 424 | 73.23% |
| YTHDF2 | 201 | 2 | 220 | 145 | 11 | 378 | 65.28% | |
| YTHDF3 | 227 | 1 | 64 | 243 | 44 | 352 | 60.79% | |
| Others | TP53 | 95 | 2 | 424 | 50 | 8 | 484 | 83.59% |
CNV, Copy Number Variant.
The Relationship Between CNVs of m6A Regulatory Genes and Clinicopathological Parameters in OC
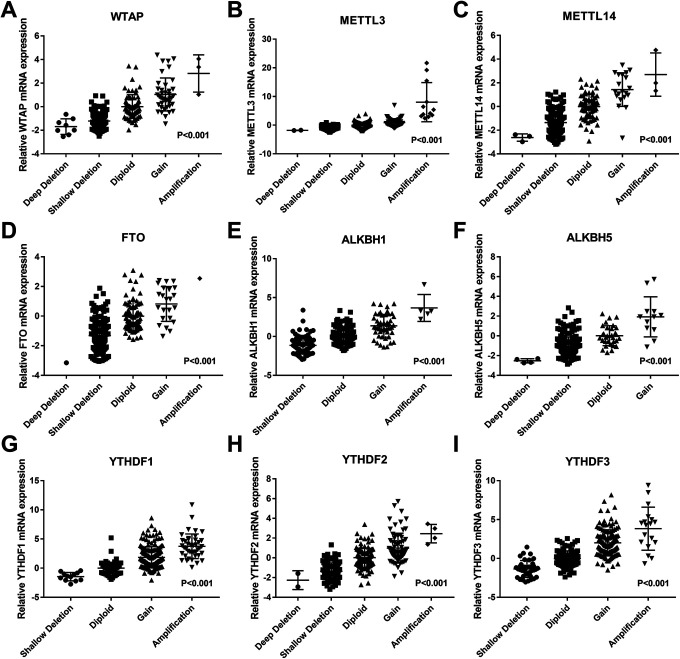
As previously mentioned, OC patients had extremely high frequency CNVs of m6A regulatory genes, and CNVs concurrence in m6A regulatory genes was a common phenomenon. To assess CNVs’ significance in OC, we explored the clinical pathological and molecular parameters in 2 groups of OC patients with different coexisting numbers of CNV genes (≤4 m6A related CNV genes and >4 m6A related CNV genes). Results showed a significant association between more coexisting m6A numbers related CNV genes with higher histologic grade and TP53 alteration. However, other parameters revealed no statistical significance between the groups with regard to age, stage, and primary tumor site (Table 2). We then explored CNVs’ significance at the transcriptional level in 307 OC samples with RNA sequencing data, and found a significant positive correlation between mRNA expression and CNV expression patterns of m6A related genes. In all 9 genes, we found higher and lower mRNA expression levels in gain and loss of copy number variations, respectively (Figure 2).
Table 2.
Clinical Pathological and Molecular Parameters of Ovarian Cancer Patients With Different Numbers of m6A Related CNV Genes.
| With CNV genes (≤4) | With CNV genes (>4) | P | ||
|---|---|---|---|---|
| Age | ≤60 | 59 | 255 | |
| >60 | 34 | 220 | 0.088 | |
| Stage | Ⅰ + Ⅱ | 8 | 39 | |
| Ⅲ | 68 | 365 | ||
| Ⅳ | 17 | 67 | 0.588 | |
| NA | 0 | 4 | ||
| Historical Grade | G1 + G2 | 20 | 53 | |
| G3 + G4 | 71 | 409 | 0.010 | |
| GX | 1 | 9 | ||
| GB | 1 | 1 | ||
| NA | 0 | 3 | ||
| Primary Tumor Site | Bilateral | 25 | 123 | |
| Left or right | 64 | 323 | 0.898 | |
| NA | 4 | 29 | ||
| TP53 | No alteration | 10 | 28 | |
| Alteration | 34 | 239 | 0.043 | |
| Not profiled | 49 | 208 |
CNV, Copy Number Variant. M6A, N6-Methyladenosine.
Figure 2.
Relationship between diverse CNV patterns and mRNA expression for m6A regulatory genes in ovarian cancer. Copy number gains or amplification had a higher mRNA expression, but deep deletions or shallow deletions showed a lower mRNA expression in m6A regulatory genes (A-I). CNV, copy number variant. m6A, N6-methyladenosine.
Prognostic Values of m6A Regulatory Genes in OC
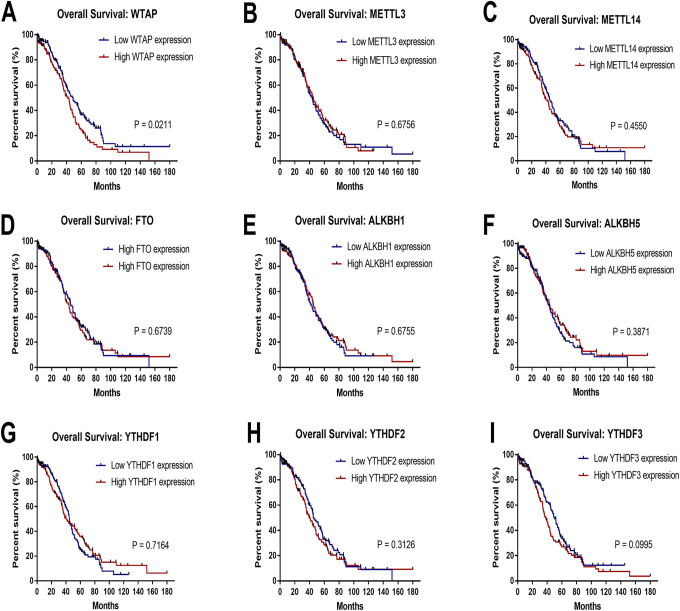
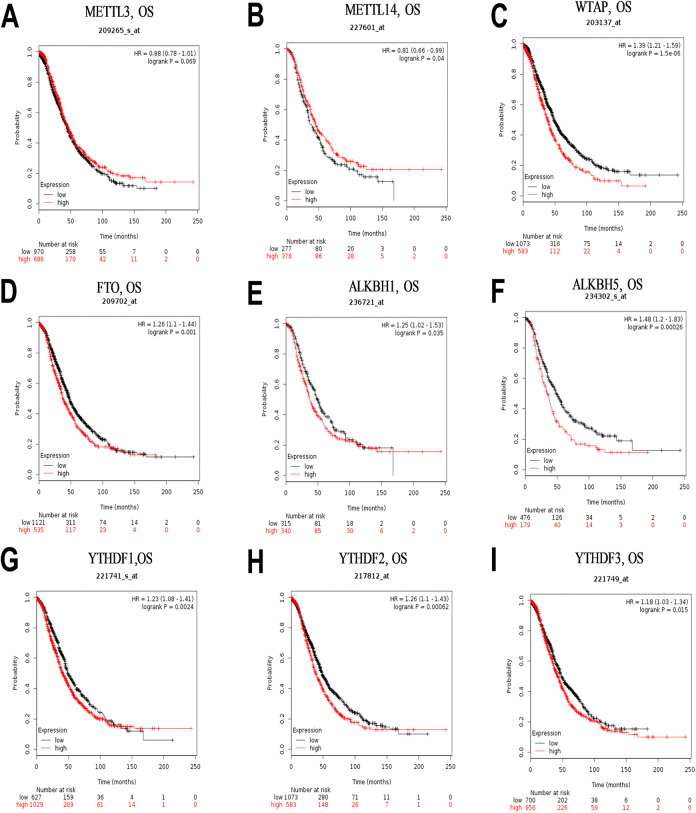
We investigated whether different CNVs patterns in m6A regulatory genes were correlated with prognosis of OC patients, and found no significant prognostic values (Supplementary Information 2). We then explored the relationship between profiles of mRNA expression across all 9 m6A regulatory genes and prognosis in OC patients using the TCGA database. Notably, high WTAP expression level was significantly correlated with the worst overall survival (OS) (p = 0.021), whereas expression patterns of the other m6A regulatory genes had no effect on OC (Figure 3). Furthermore, we used the online Kaplan-Meier plotter database to examine the prognostic value of m6A regulatory genes in OC, and found an association between high mRNA expression levels of WTAP, FTO, ALKBH1, ALKBH5, YTHDF1, YTHDF2, and YTHDF3, as well as low levels of METTL14, with poor prognosis (Figure 4). Conversely, METTL3 was not associated with prognosis in ovarian cancer patients. Although the high and low levels of m6A regulatory gene expression demonstrated distinct prognostic value depending on the different databases, it was evident that WTAP is a potential prognostic factor for OC.
Figure 3.
Relationship between different patterns of mRNA expression for m6A regulatory genes and prognosis in ovarian cancer. High mRNA expression levels of WTAP(A) was associated with worse OS, while METTL3(B), METTL14(C), FTO(D), ALKBH1(E), ALKBH5(F), YTHDF1(G), YTHDF2(H), and YTHDF3(I) had no significant effect on OS in the TCGA cohort. OS, overall survival. m6A, N6-methyladenosine.
Figure 4.
Kaplan–Meier survival curves for m6A related genes expression in ovarian cancer (A–I). High mRNA expression levels of WTAP (C), FTO (D), ALKBH1 (E), ALKBH5 (F), YTHDF1 (G), YTHDF2 (H), and YTHDF3 (I) and low mRNA expression levels of METTL14 (B) were associated with worse OS, whereas METTL3 (A) had no significant effect on OS in the KM cohort. OS, overall survival. m6A, N6-methyladenosine.
The Relationship Between Clinicopathological Characteristics and Gene Expression With Survival
We used the multivariate Cox proportional hazards model to explore the effect of m6A regulatory gene expression and clinicopathological characteristics on survival. Results showed that age (p = 0.027) and WTAP (p = 0.024) mRNA expression were independent prognostic factors (Table 3), whereas race, stage, grade, as well as expression of METTL3, METTL14, FTO, ALKBH1, ALKBH5, YTHDF1, YTHDF2, and YTHDF3 were not independent prognostic factors in OC.
Table 3.
Effect of Expression Profiles of m6A Regulatory Genes and Clinicopathological Characteristics on Survival Based on Multivariate Cox Proportional Hazards Model.
| Ovarian cancer (N = 231) | Variable | Coef | HR | 95%CI_l | 95%CI_u | P-value |
|---|---|---|---|---|---|---|
| Age | 0.364 | 1.439 | 1.042 | 1.988 | 0.027 | |
| Race | -0.223 | 0.800 | 0.465 | 1.377 | 0.421 | |
| Stage | 0.335 | 1.397 | 0.632 | 3.090 | 0.409 | |
| Grade | 0.423 | 1.527 | 0.934 | 2.497 | 0.092 | |
| WTAP | 0.174 | 1.191 | 1.023 | 1.385 | 0.024 | |
| METTL3 | 0.046 | 1.047 | 0.874 | 1.255 | 0.618 | |
| METTL14 | -0.001 | 0.999 | 0.851 | 1.173 | 0.995 | |
| FTO | 0.149 | 1.161 | 0.991 | 1.361 | 0.065 | |
| ALKBH1 | -0.091 | 0.913 | 0.770 | 1.082 | 0.293 | |
| ALKBH5 | -0.035 | 0.965 | 0.831 | 1.121 | 0.642 | |
| YTHDF1 | -0.011 | 0.989 | 0.837 | 1.170 | 0.900 | |
| YTHDF2 | 0.056 | 1.058 | 0.898 | 1.246 | 0.504 | |
| YTHDF3 | 0.045 | 1.046 | 0.900 | 1.215 | 0.561 |
M6A, N6-Methyladenosine. Coef, Regression Coefficient; HR, Hazard Ratio; 95%CI_l, 95% Confidence Interval Lower Limit; 95%CI_u, 95% Confidence Interval Upper Limit.
Levels of WTAP Expression in OC
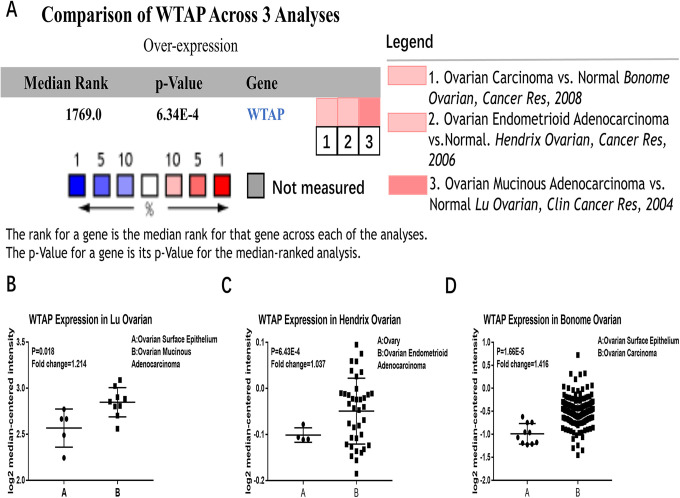
Screening of the Oncomine microarray database revealed significantly higher RNA levels of WTAP in OC tissues than healthy controls across 3 representative datasets (Figure 5A), including the Lu, Hendrix, and Bonome Ovarian dataset29-31 (Figure 5B-5D).
Figure 5.
Expression of WTAP in the Oncomine database. (A) Comparisons of the expression of WTAP in OC in 3 independent analyses from the Oncomine database. Validation of WTAP expression in ovarian cancer in the Lu Ovarian dataset (B), Hendrix Ovarian dataset (C), and Bonome Ovarian dataset (D). (*P < 0.05; **P < 0.01; ***P < 0.001).
Enrichment Analysis of the WTAP Gene
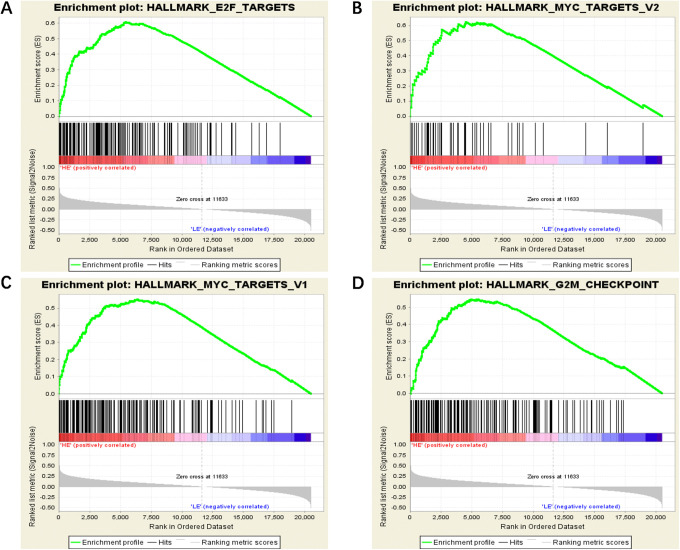
Considering WTAP’s vital role in regulating a variety of crucial biological functions (especially during the m6A modification of mRNA), we determined the biological processes in which this factor is involved in OC. Firstly, we divided patterns of WTAP’s RNA expression into 2 groups, then performed Gene set enrichment analysis (GSEA). Finally, we identified 4 significant high-scoring sets, including E2F_TARGETS, MYC_TARGETS_V2, MYC_TARGETS_V1, and G2M_CHECKPOINT (see Table 4 and Figure 6). These results provide new insights into the role of epigenetic regulation in OC.
Table 4.
Four High-Scoring Sets are Identified by GSEA Analysis.
| GS DETAILS | SIZE | ES | NES | NOM p-val | FDR q-val |
|---|---|---|---|---|---|
| HALLMARK_E2F_TARGETS | 162 | 0.61 | 1.83 | 0.018 | 0.082 |
| HALLMARK_MYC_TARGETS_V2 | 50 | 0.62 | 1.81 | 0.006 | 0.043 |
| HALLMARK_MYC_TARGETS_V1 | 165 | 0.55 | 1.78 | 0.016 | 0.039 |
| HALLMARK_G2M_CHECKPOINT | 158 | 0.55 | 1.73 | 0.032 | 0.044 |
GSEA, Gene Set Enrichment Analysis.
Figure 6.
GSEA results for WTAP in OC patients. Four high-scoring sets, including E2F TARGETS (A), MYC TARGETS V2 (B), MYC TARGETS V1(C), and G2M CHECKPOINT (D), are identified by GSEA analysis. GSEA, Gene set enrichment analysis.
Discussion
Generally, m6A writers, erasers, and readers might show diverse patterns across distinct malignancies or independent databases. Results from the present study have shown that a higher frequency of both CNVs in single m6A related genes as well as concurrent CNVs in 2 or more genes, relative to those previously reported in kidney malignancy27 and AML.28 To some extent, our findings suggest that regulation disorder of m6A might have a more significant impact on development and progression of ovarian cancer. Previous studies have demonstrated the synergistic roles played by multiple m6A related genes in formation of complexes.5 In the present study, it was evident that WTAP, the m6A “writer” gene, was more predisposed to CNVs and had a significantly higher prognostic value than other m6A related genes in OC. However, other genes, such as METTL3, FTO, and ALKBH5, have been previously shown to be more crucial in kidney malignancy,27 breast cancer,18 glioblastoma,15 and AML.32 Analysis of the TCGA databases revealed that only high WTAP expression was significantly associated with more inferior OS. Additionally, results from the Kaplan-Meier plotter database also showed that high expression of WTAP, FTO, ALKBH1, ALKBH5, YTHDF1, YTHDF2, and YTHDF3, as well as low expression of METTL14, were all associated with poor OS. These differences might be attributed to the diverse methods of data detection and analysis as well as different mechanisms of inherent biological characteristics. Despite the inconsistency, we still found a consistent prognostic correlation between WTAP mRNA expression and OC, based on an integrative bioinformatics analysis. The resulting diverse expression patterns of m6A regulatory genes across different tumor types or in independent databases affirmed the complexity of the m6A’s post-transcriptional regulation mechanism, and suggested presence of tumor specificity of the m6A regulators.
WTAP, located in the nucleus, was first identified as a partner of Wilms’ tumor 1 protein.33 It was then mapped to human chromosome 6q25-27, and an allele loss detected in all histological OC types.34 Interestingly, our research also found that a shallow deletion in the WTAP gene was the high-frequency pattern of CNVs. Previous studies have described WTAP in the context of cell proliferation and apoptosis of vascular smooth muscle cells before.35-37 Recently, the gene was found to play a crucial role in the tumorigenesis of various malignancies, such as AML, cholangiocarcinoma, endometrial cancer, and glioblastoma.38-41 Besides, accumulating evidence suggests that WTAP is a new constituent of the human m6A multiprotein writer complex, and plays a role in facilitating recruitment of the m6A complex to a target site.42 In fact, some studies have demonstrated that the WTAP complex plays a crucial role in cell cycle regulation, including G2/M transition, by stabilizing cyclin A2 mRNA43 and CDK2 mRNA,44,45 which is consistent with our hypothesis. Wilms tumor 1 gene, a partner of WTAP, has also been suggested to be an oncogene that induces expression of MYC.46,47 Besides, previous studies have suggested that some key transcription factors, such as FOXO1 and IRF1, might be involved in regulation of WTAP promoter, and are also be closely related to tumorigenesis, tumor growth, and invasion.48 WTAP has also been found to be highly expressed in ovarian cancer49,50 and associated with worse survival outcomes in high-grade serous ovarian carcinoma.49 However, this evidence is still insufficient, while the underlying molecular mechanism of WTAP in OC remains unknown.
Our study had some limitations. Firstly, this was a retrospective analysis, based on publicly available popular datasets. Therefore, the number of included patients was limited, and the results may not be as reliable as those from prospective studies. Secondly, more in vivo and in vitro experiments are required for functional and clinical validation.
In some ways, the function of m6A RNA methylation regulators in tumorigenesis may be confusing. For instance, WTAP is reportedly an overexpressed oncogene, with its high expression strongly correlated with poor survival in bladder cancer,51 renal cell carcinoma,44 pancreatic ductal adenocarcinoma,52 and Malignant Glioma,53 which corroborates our findings from OC. Conversely, we found that shallow deletion was WTAP’s main CNV pattern, with loss of copy number events significantly associated with lower WTAP mRNA expression. This contradiction is puzzling and may be attributed to the distinctively dynamic process of m6A regulation or the heterogeneity of the tumor. This affirms WTAP’s function as a multi-dimensional oncogene.54
Conclusion
Overall our findings demonstrate the significance of high-frequency genetic alterations of m6A RNA methylation regulators and WTAP’s poor prognosis value in OC. These findings provide new insights into the role of m6A methylation in OC, and will be vital in guiding development of novel treatment therapies.
Supplemental Material
Supplemental Material, Supplementary_Information_1 for Gene Signatures and Prognostic Values of m6A RNA Methylation Regulators in Ovarian Cancer by Xiao Han, Jie Liu, Guomei Cheng and Shihong Cui in Cancer Control
Supplemental Material, Supplementary_Information_2 for Gene Signatures and Prognostic Values of m6A RNA Methylation Regulators in Ovarian Cancer by Xiao Han, Jie Liu, Guomei Cheng and Shihong Cui in Cancer Control
Footnotes
Authors’ Note: All patient-related raw data was downloaded from open-access public databases and it is confirmed that all participants involved have gave written informed consent.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by Grants-in-Aid from the Medical Innovation Talent Program, Health Department of Henan Province, China (Project No. 201702106).
ORCID iD: Shihong Cui  https://orcid.org/0000-0002-6691-1646
https://orcid.org/0000-0002-6691-1646
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2. Prat J; FIGO Committee on Gynecologic Oncology. Staging classification for cancer of the ovary, fallopian tube, and peritoneum. Int J Gynaecol Obstet. 2014;124(1):1–5. doi:10.1016/j.ijgo.2013.10.001 [DOI] [PubMed] [Google Scholar]
- 3. Liu N, Pan T. N6-methyladenosine-encoded epitranscriptomics. Nat Struct Mol Biol. 2016;23(2):98–102. doi:10.1038/nsmb.3162 [DOI] [PubMed] [Google Scholar]
- 4. Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18(1):31–42. doi:10.1038/nrm.2016.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Deng X, Su R, Weng H, Huang H, Li Z, Chen J. RNA N(6)-Methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018;28(5):507–517. doi:10.1038/s41422-018-0034-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Du K, Zhang L, Lee T, Sun T. m(6)A RNA Methylation controls neural development and is involved in human diseases. Mol Neurobiol. 2019;56(3):1596–1606. doi:10.1007/s12035-018-1138 -1 [DOI] [PubMed] [Google Scholar]
- 7. Engel M, Eggert C, Kaplick PM, et al. The Role of m(6)A/m-RNA methylation in stress response regulation. Neuron. 2018;99(2):389–403. e389. doi:10.1016/j.neuron.2018.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mendel M, Chen KM, Homolka D, et al. Methylation of structured RNA by the m(6)A writer METTL16 is essential for mouse embryonic development. Mol Cell. 2018;71(6):986–1000. e1011 doi:10.1016/j.molcel.2018.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yoon KJ, Ringeling FR, Vissers C, et al. Temporal control of mammalian cortical neurogenesis by m(6)a methylation. Cell. 2017;171(4):877–889. e817. doi:10.1016/j.cell.2017.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang X, He C. Reading RNA methylation codes through methyl-specific binding proteins. RNA Biol. 2014;11(6):669–672. doi:10.4161/rna.28829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ma JZ, Yang F, Zhou CC, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65(2):529–543. doi:10.1002/hep.28885 [DOI] [PubMed] [Google Scholar]
- 12. Yang Z, Li J, Feng G, et al. MicroRNA-145 modulates N(6)-Methyladenosine levels by targeting the 3’-untranslated mRNA region of the N(6)-Methyladenosine binding YTH domain family 2 protein. J Biol Chem. 2017;292(9):3614–3623. doi:10.1074/jbc.M116.749689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao X, Chen Y, Mao Q, et al. Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomark. 2018;21(4):859–868. doi:10.3233/CBM-170791 [DOI] [PubMed] [Google Scholar]
- 14. Cui Q, Shi H, Ye P, et al. m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18(11):2622–2634. doi:10.1016/j.celrep.2017.02.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang S, Zhao BS, Zhou A, et al. m(6)A Demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31(4):591–606. e596 doi:10.1016/j.ccell.2017.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weng H, Huang H, Wu H, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell. 2018;22(2):191–205. e199 doi:10.1016/j.stem.2017.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang C, Samanta D, Lu H, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc Natl Acad Sci U S A. 2016;113(14):E2047–E2056. doi:10.1073/pnas.1602883113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang C, Zhi WI, Lu H, et al. Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217- and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget. 2016;7(40):64527–64542. doi:10.18632/oncotarget.11743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu J, Ren D, Du Z, Wang H, Zhang H, Jin Y. m(6)A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem Biophys Res Commun. 2018;502(4):456–464. doi:10.1016/j.bbrc.2018.05.175 [DOI] [PubMed] [Google Scholar]
- 20. Xu D, Shao W, Jiang Y, Wang X, Liu Y, Liu X. FTO expression is associated with the occurrence of gastric cancer and prognosis. Oncol Rep. 2017;38(4):2285–2292. doi:10.3892/or.2017.5904 [DOI] [PubMed] [Google Scholar]
- 21. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1 doi:10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–180. doi:10.1593/neo.07112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lanczky A, Nagy Á, Bottai G, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160(3):439–446. doi:10.1007/s10549-016-4013-7 [DOI] [PubMed] [Google Scholar]
- 24. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi:10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi:10.1093/bioinformatics/btr260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mullany LK, Wong KK, Marciano DC, et al. Specific TP53 mutants overrepresented in ovarian cancer impact CNV, TP53 activity, responses to nutlin-3a, and cell survival. Neoplasia. 2015;17(10):789–803. doi:10.1016/j.neo.2015.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou J, Wang J, Hong B, et al. Gene signatures and prognostic values of m6A regulators in clear cell renal cell carcinoma – a retrospective study using TCGA database. Aging (Albany NY). 2019;11(6):1633–1647. doi:10.18632/aging.101856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kwok CT, Marshall AD, Rasko JE, Wong JJ. Genetic alterations of m(6)A regulators predict poorer survival in acute myeloid leukemia. J Hematol Oncol. 2017;10(1):39 doi:10.1186/s13045-017-0410-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bonome T, Levine DA, Shih J, et al. A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res. 2008;68(13):5478–5486. doi:10.1158/0008-5472.CAN-07-6595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hendrix ND, Wu R, Kuick R, Schwartz DR, Fearon ER, Cho KR. Fibroblast growth factor 9 has oncogenic activity and is a downstream target of Wnt signaling in ovarian endometrioid adenocarcinomas. Cancer Res. 2006;66(3):1354–1362. doi:10.1158/0008-5472.CAN-05-3694 [DOI] [PubMed] [Google Scholar]
- 31. Lu KH, Patterson AP, Wang L, et al. Selection of potential markers for epithelial ovarian cancer with gene expression arrays and recursive descent partition analysis. Clin Cancer Res. 2004;10(10):3291–3300. doi:10.1158/1078-0432.CCR-03-0409 [DOI] [PubMed] [Google Scholar]
- 32. Li Z, Weng H, Su R, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-Methyladenosine RNA Demethylase. Cancer Cell. 2017;31(1):127–141. doi:10.1016/j.ccell.2016.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Little NA, Hastie ND, Davies RC. Identification of WTAP, a novel Wilms’ tumour 1-associating protein. Hum Mol Genet. 2000;9(15):2231–2239. doi:10.1093/oxfordjournals.hmg.a018914 [DOI] [PubMed] [Google Scholar]
- 34. Cooke IE, Shelling AN, Le Meuth VG, Charnock ML, Ganesan TS. Allele loss on chromosome arm 6q and fine mapping of the region at 6q27 in epithelial ovarian cancer. Genes Chromosomes Cancer. 1996;15(4):223–233. doi:10.1002/(SICI)1098-2264(199604)15:4<223:: AID-GCC4>3.0.CO;2-3 [DOI] [PubMed] [Google Scholar]
- 35. Small TW, Pickering JG. Nuclear degradation of Wilms tumor 1-associating protein and survivin splice variant switching underlie IGF-1-mediated survival. J Biol Chem. 2009;284(37):24684–24695. doi:10.1074/jbc.M109.034629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Small TW, Penalva LO, Pickering JG. Vascular biology and the sex of flies: regulation of vascular smooth muscle cell proliferation by Wilms’ tumor 1-associating protein. Trends Cardiovasc Med. 2007;17(7):230–234. doi:10.1016/j.tcm.2007.08.002 [DOI] [PubMed] [Google Scholar]
- 37. Small TW, Bolender Z, Bueno C, et al. Wilms’ tumor 1-associating protein regulates the proliferation of vascular smooth muscle cells. Circ Res. 2006;99(12):1338–1346. doi:10.1161/01.RES.0000252289.79841.d3 [DOI] [PubMed] [Google Scholar]
- 38. Jin DI, Lee SW, Han ME, et al. Expression and roles of Wilms’ tumor 1-associating protein in glioblastoma. Cancer Sci. 2012;103(12):2102–2109. doi:10.1111/cas.12022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bansal H, Yihua Q, Iyer SP, et al. WTAP is a novel oncogenic protein in acute myeloid leukemia. Leukemia. 2014;28(5):1171–1174. doi:10.1038/leu.2014.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jo HJ, Shim HE, Han ME, et al. WTAP regulates migration and invasion of cholangiocarcinoma cells. J Gastroenterol. 2013;48(11):1271–1282. doi:10.1007/s00535-013-0748-7 [DOI] [PubMed] [Google Scholar]
- 41. Liu J, Eckert MA, Harada BT, et al. m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol. 2018;20(9):1074–1083. doi:10.1038/s41556-018-0174-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ping XL, Sun BF, Wang L, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24(2):177–189. doi:10.1038/cr.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Horiuchi K, Umetani M, Minami T, et al. Wilms’ tumor 1-associating protein regulates G2/M transition through stabilization of cyclin A2 mRNA. Proc Natl Acad Sci U S A. 2006;103(46):17278–17283. doi:10.1073/pnas.0608357103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tang J, Wang F, Cheng G, et al. Wilms’ tumor 1-associating protein promotes renal cell carcinoma proliferation by regulating CDK2 mRNA stability. J Exp Clin Cancer Res. 2018;37(1):40 doi:10.1186/s13046-018-0706-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Horiuchi K, Kawamura T, Iwanari H, et al. Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem. 2013;288(46):33292–33302. doi:10.1074/jbc.M113.500397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ariyaratana S, Loeb DM. The role of the Wilms tumour gene (WT1) in normal and malignant haematopoiesis. Expert Rev Mol Med. 2007;9(14):1–17. doi:10.1017/S1462399407000336 [DOI] [PubMed] [Google Scholar]
- 47. Rather MI, Swamy S, Gopinath KS, Kumar A. Transcriptional repression of tumor suppressor CDC73, encoding an RNA polymerase II interactor, by Wilms tumor 1 protein (WT1) promotes cell proliferation: implication for cancer therapeutics. J Biol Chem. 2014;289(2):968–976. doi:10.1074/jbc.M113.483255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu LS, Qian JY, Wang M, Yang H. Identifying the role of Wilms tumor 1 associated protein in cancer prediction using integrative genomic analyses. Mol Med Rep. 2016;14(3):2823–2831. doi:10.3892/mmr.2016.5528 [DOI] [PubMed] [Google Scholar]
- 49. Yu HL, Ma XD, Tong JF, Li JQ, Guan XJ, Yang JH. WTAP is a prognostic marker of high-grade serous ovarian cancer and regulates the progression of ovarian cancer cells. Onco Targets Ther. 2019;12:6191–6201. doi:10.2147/OTT.S205730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sommer A, Hilpert F, Arnold N. Gene expression profiling of epithelial ovarian cancer. Curr Genomics. 2006;7(2):115–135. doi:10.2174/138920206777304641 [Google Scholar]
- 51. Chen L, Wang X. Relationship between the genetic expression of WTAP and bladder cancer and patient prognosis. Oncol Lett. 2018;16(6):6966–6970. doi:10.3892/ol.2018.9554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li BQ, Huang S, Shao QQ, et al. WT1-associated protein is a novel prognostic factor in pancreatic ductal adenocarcinoma. Oncol Lett. 2017;13(4):2531–2538. doi:10.3892/ol.2017.5784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xi Z, Xue Y, Zheng J, Liu X, Ma J, Liu Y. WTAP Expression predicts poor prognosis in malignant glioma patients. J Mol Neurosci. 2016;60(2):131–136. doi:10.1007/s12031-016-0788-6 [DOI] [PubMed] [Google Scholar]
- 54. Wang S, Chai P, Jia R, Jia R. Novel insights on m(6)A RNA methylation in tumorigenesis: a double-edged sword. Mol Cancer. 2018;17(1):101 doi:10.1186/s12943-018-0847-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, Supplementary_Information_1 for Gene Signatures and Prognostic Values of m6A RNA Methylation Regulators in Ovarian Cancer by Xiao Han, Jie Liu, Guomei Cheng and Shihong Cui in Cancer Control
Supplemental Material, Supplementary_Information_2 for Gene Signatures and Prognostic Values of m6A RNA Methylation Regulators in Ovarian Cancer by Xiao Han, Jie Liu, Guomei Cheng and Shihong Cui in Cancer Control