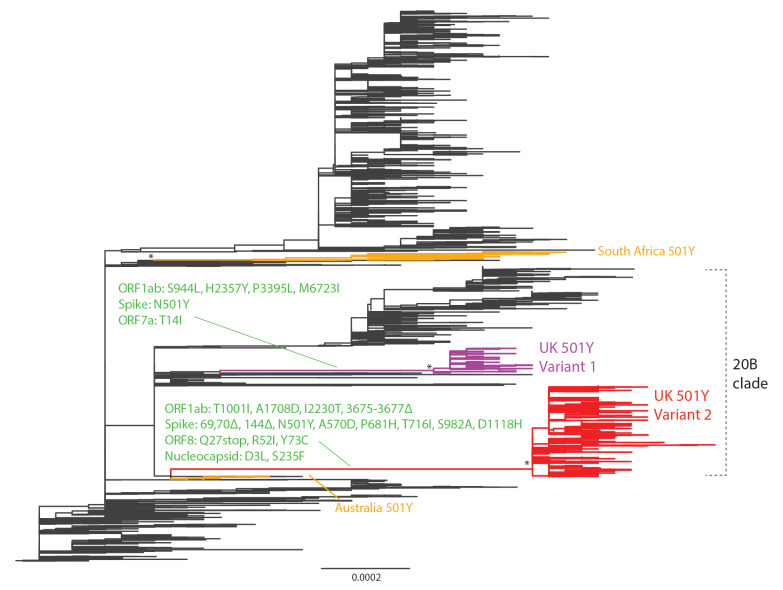
Figure 1.
Phylogeny of SARS-CoV-2 showing the emergence of 501Y lineages, United Kingdom and other regions, as at 19 December 2020
Nt: nucleotide; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2; UK: United Kingdom; US: United States.
The maximum likelihood tree was built from 7,003 genome nt sequences of SARS-CoV-2. The UK 501Y Variant 1 and 2 lineages are coloured in purple and red, respectively. The South African and Australian 501Y lineages are in orange. Amino acid changes at the preceding branches of UK 501Y Variant 1 and 2 are indicated in green. Some 501Y variants in sporadic emergence (including those in Spain and the US, etc.) without establishment of a lineage of more than 10 sequences are not shown. The asterisks refer to the > 0.98 topology supports (Shimodaira-Hasegawa test) for the monophyletic grouping of the 501Y lineages. Branch scale is shown at the bottom of the tree in the unit of substitutions per site.