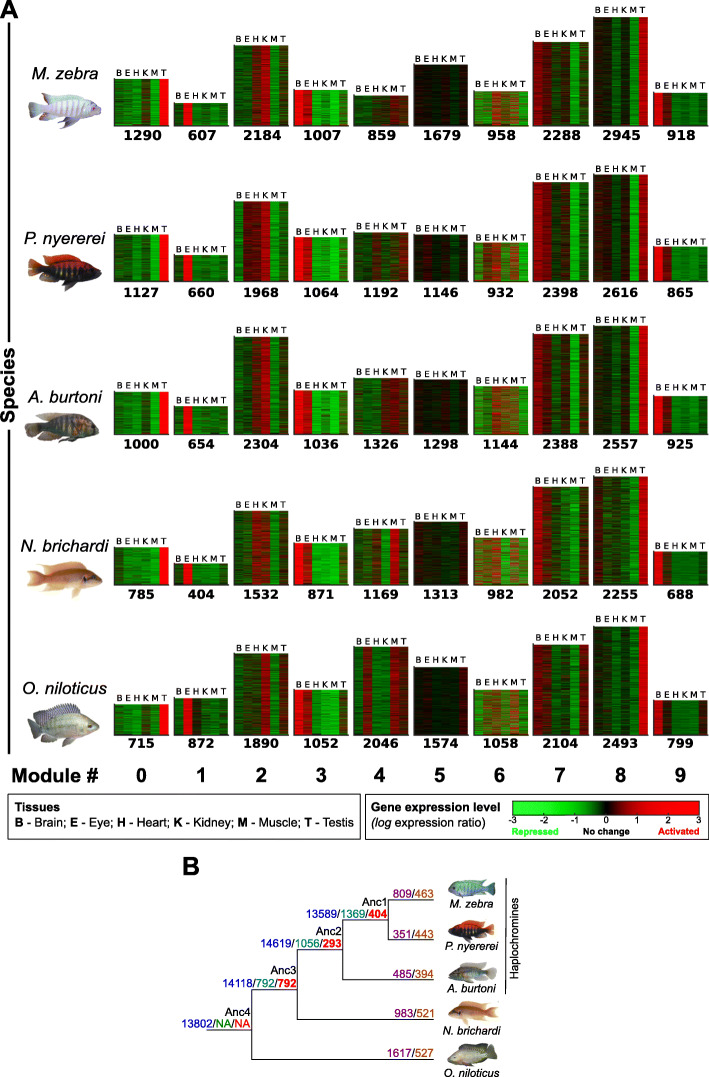
Fig. 1.
Evolution of gene expression in five cichlids. a Ten (0–9, heatmaps) co-expression modules identified by Arboretum [9] in six tissues of five cichlid species. Color bar denotes log expression ratio across each tissue, relative to the mean expression across all tissues—(red) activated, (green) repressed and (black) no change. Each heatmap shows the expression profile of genes assigned to that module in a given species and height is proportional to number of genes in the module (on bottom). b Number of state changes in module assignment of 1-to-1 orthologous genes along the five cichlid phylogeny [18]. Blue numbers: ancestral node genes assigned to modules; green numbers: state changes compared to the deepest common ancestor (Anc4); red numbers: state changes from last common ancestor (LCA); purple numbers: state changes in the one “focal” species compared to all other species; orange numbers: convergent state changes in each of the “focal” species and any of the other species