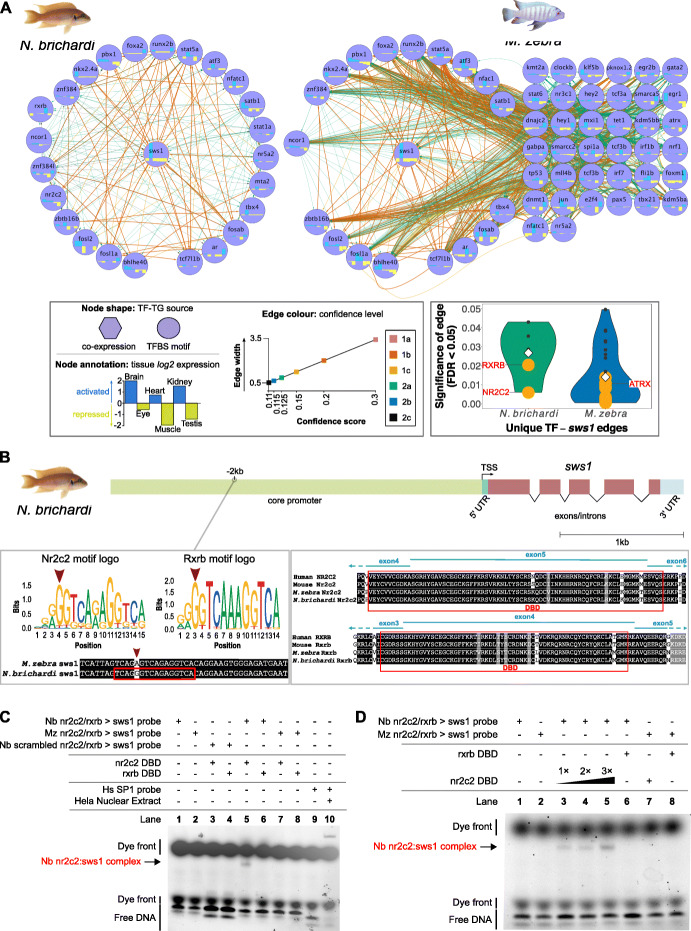
Fig. 3.
Evolution of the sws1 opsin regulatory networks in N. brichardi and M. zebra. a Reconstructed regulatory networks of sws1 opsin shown for N. brichardi (left) and M. zebra (right): circular layout nodes are common regulators (unless missing); grid layout nodes are unique regulators in M. zebra. Node shape, annotation and edge color denoted in legend to left bottom. Violin plot of significance (FDR < 0.05) of unique TF-sws1 edges in N. brichardi (green violin) and M. zebra (blue violin) to bottom right—mean edge significance score shown within each plot (white diamond); edges more than the mean (less significant) are shown as gray dots, and edges less than the mean (more significant) are shown as orange dots; selected example TFs are demarcated within. b On the left, NR2C2 and RXRB motif logos and motif prediction in negative orientation N. brichardi sws1 gene promoter (red box) and variant in M. zebra sws1 gene promoter (red arrow). On the right, NR2C2 and RXRB partial protein alignment showing DNA-binding domain (DBD) annotation in human, mouse, M. zebra and N. brichardi. c EMSA validation of NR2C2 and RXRB DBD binding to N. brichardi and M. zebra sws1 gene promoter. Table denotes combinations of DNA probe and expressed DBD in EMSA reactions that include negative controls (lanes 1 to 4); N. brichardi protein: DNA-binding assay (lanes 5 and 6); M. zebra protein: DNA-binding assay (lanes 7 and 8); kit negative (lane 9) and binding positive control (lane 10). Protein:DNA complexes, dye front and free DNA are indicated by arrowhead and bracket within. d EMSA validation of increasing NR2C2 DBD concentrations and binding to predicted TFBS in N. brichardi sws1 gene promoter