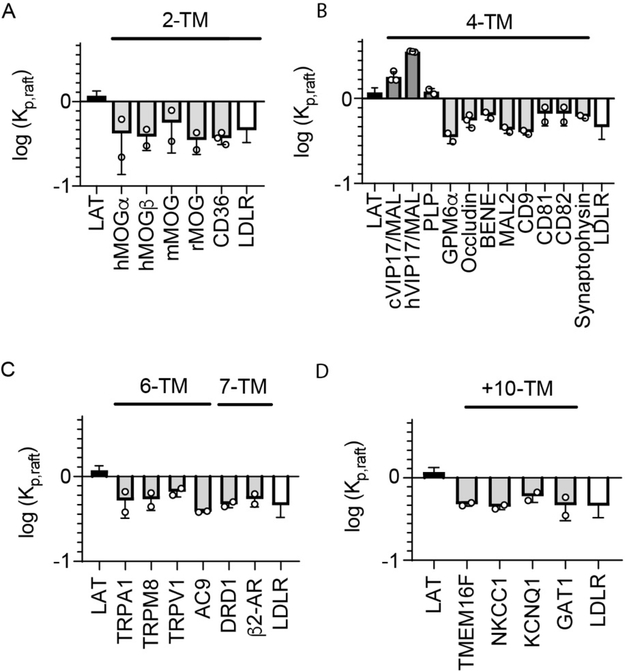
Figure 2 – Almost all multi-pass TMPs have low raft affinity in GPMVs.
Shown are the raft affinity of multi-pass TMPs with varying number of TMHs. Raft affinity is shown in log scale, with 0 representing equal partitioning between the phases and positive and negative values representing enrichment in or depletion from the raft phase, respectively. Included in each panel is the value for LAT (a raft-preferring single pass TMD, black) and LDLR (a non-raft TMD, white). (A) 2-TM proteins. Three homologs of MOG from different species (h:human; m:mouse; r:rat) and both isoforms (alpha and beta) of the human protein are excluded from rafts, as is the unrelated CD36. (B) Two 4-TM proteins are highly raft-preferring, namely MAL and PLP. In contrast, several other members of the MARVEL family (BENE, MAL2, occludin, and synaptophysin) and other tetraspanin family proteins (CD9, CD81, CD82), as well as the neuronal membrane glycoprotein (GPM6a) are almost completely excluded. Two analogs of MAL (c:canine: h:human) were tested and both had significantly greater raft affinity than LAT. (C) All tested 6-TM and 7-TM proteins are raft excluded. Several proteins from the TRP family (TRPA1, TRPM8, TRPV1), Adenylyl Cyclase 9 (AC9), and G Protein-Coupled Receptors (DRD1, β2AR) were not significantly different from LDLR. (D) All tested proteins with 10 or more TMDs - PM scramblase TMEM16F and several transporters (NKCC1, KCNQ1 and GAT1) - were raft excluded. Data points represent the means of individual experiments of >10 vesicles each.