Fig. 5.
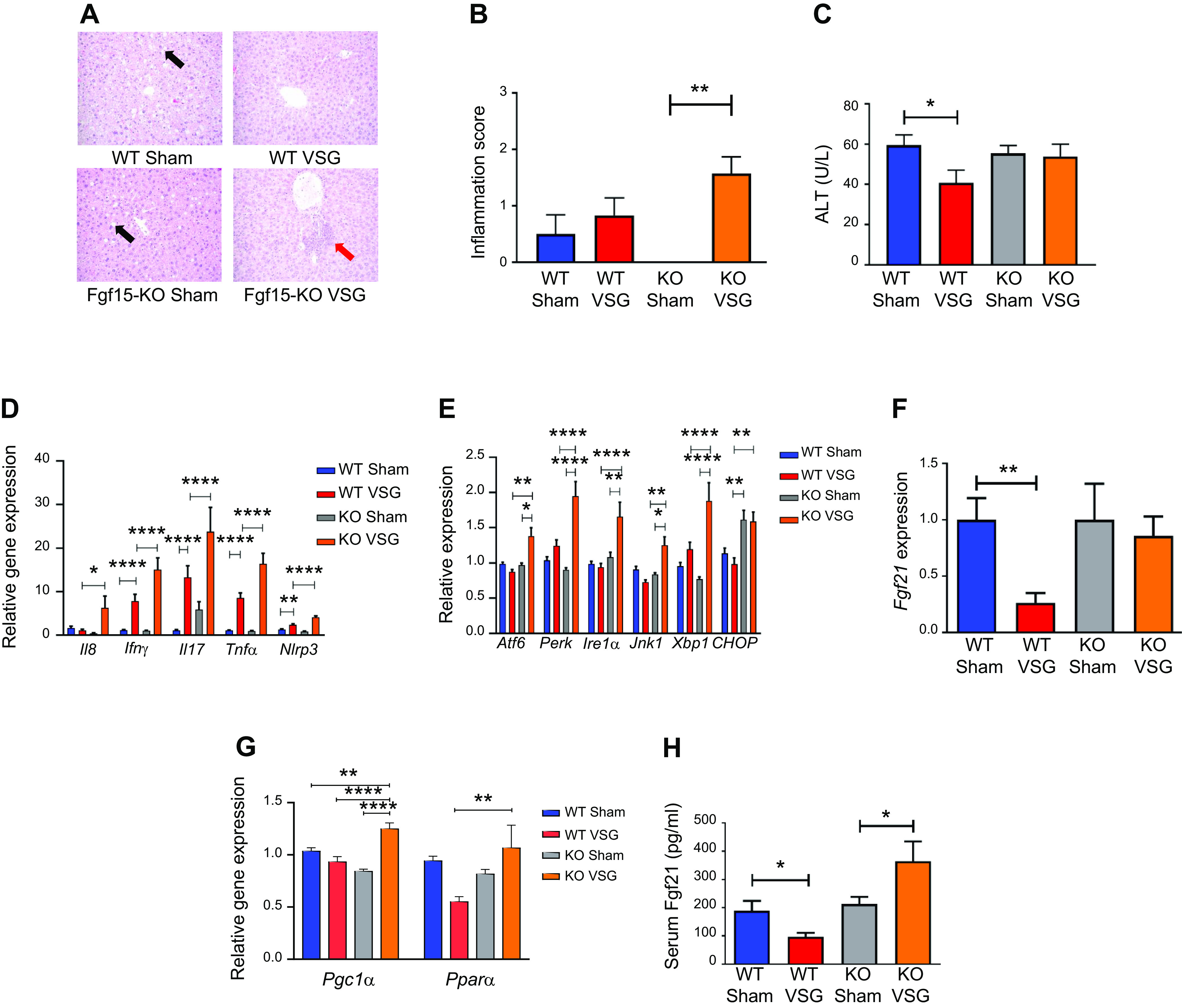
Fibroblast growth factor 15 (Fgf15) deficiency results in increased hepatic inflammation via an endoplasmic reticulum (ER) stress mechanism post-post-vertical sleeve gastrectomy (VSG). A: hepatic histology 49 days postsurgery. Representative ×20 magnified hematoxylin-eosin sections showed lipid droplet accumulation in livers of Sham-operated groups (black arrow), whereas this was not observed in VSG groups. In knockout (KO) VSG mice, we observed inflammation foci in liver parenchyma (red arrow). B: Hepatic inflammation score 49 days postsurgery. KO VSG mice developed inflammation in liver compared with their control group, which is expressed quantitatively by inflammation score bar graph. No inflammation was observed in KO Sham liver sections; therefore, no inflammation score was plotted for same; n: wild-type (WT) Sham = 6, WT VSG = 6, KO Sham = 5, KO VSG = 7. **P < 0.01, one-way ANOVA. C: serum alanine aminotransferase (ALT) levels 49 days postsurgery. ALT was lower in WT VSG mice compared with WT Sham mice 49 days postsurgery; n: WT Sham = 6, WT VSG = 6, KO Sham = 5, KO VSG = 7. *P < 0.05, t test. D: hepatic inflammation gene expression at day 49 postsurgery. mRNA levels of genes coding for inflammation were measured by Biomark HD System high-throughput PCR and expressed in relative expression units. Interleukin-8 (Il8), interferon-γ (IFNγ), interleukin-17 (Il17), tumor necrosis factor-α (TNFα), and Nlrp3 (NOD-, LRR- and pyrin domain-containing protein-3) were significantly overexpressed only in KO VSG mice; n: WT Sham = 4, WT VSG = 5, KO Sham = 4, KO VSG = 6; *P < 0.05, **P < 0.01, ****P < 0.0001, one-way ANOVA, t test; n: WT Sham = 6, WT VSG = 6, KO Sham = 5, KO VSG = 7; (*P < 0.05, **P < 0.01, ****P < 0.0001, one-way ANOVA. E: hepatic ER stress gene expression at day 49 postsurgery. mRNA levels of genes coding for ER stress were measured by Biomark HD System high-throughput PCR and expressed in relative expression units. Atf6, Perk, Ire1a, Jnk1, and Xbp1 were significantly overexpressed only in KO VSG mice; n: WT Sham = 4, WT VSG = 5, KO Sham = 4, KO VSG = 6; *P < 0.05, **P < 0.01, ****P < 0.0001, one-way ANOVA, t test; n: WT Sham = 6, WT VSG = 6, KO Sham = 5, KO VSG = 7; *P < 0.05, **P < 0.01, ****P < 0.0001, one-way ANOVA. F: hepatic Fgf21 gene expression at day 49 postsurgery. mRNA levels of the gene coding for Fgf21 was measured by RT-PCR and expressed in relative expression units. Fgf21 was suppressed in WT VSG compared with WT Sham group, whereas this was not observed in KO VSG mice; n: WT Sham = 4, WT VSG = 5, KO Sham = 4, KO VSG = 6. **P < 0.01, one-way ANOVA, t test. G: hepatic PPARα (peroxisome proliferator-activated receptor-α) and PGC-1α (PPARγ coactivator 1α) gene expression at day 49 postsurgery. mRNA levels of PPAR.α and PGC-1α genes were measured by Biomark HD System high-throughput PCR and expressed in relative expression units. PPARα was suppressed in WT VSG compared with KO VSG group, whereas PGC-1α was overexpressed in KO VSG mice relative to all other groups; n: WT Sham = 6, WT VSG = 6, KO Sham = 5, KO VSG = 7. *P < 0.05, **P < 0.01, ****P < 0.0001, one-way ANOVA. H: serum Fgf21 levels at day 49 postsurgery. Serum Fgf21 levels were decreased in WT VSG mice, whereas they were drastically increased in KO VSG mice compared with their respective Sham controls; n: WT Sham = 6, WT VSG = 6, KO Sham = 5, KO VSG = 7. *P < 0.05, one-way ANOVA, t test.
