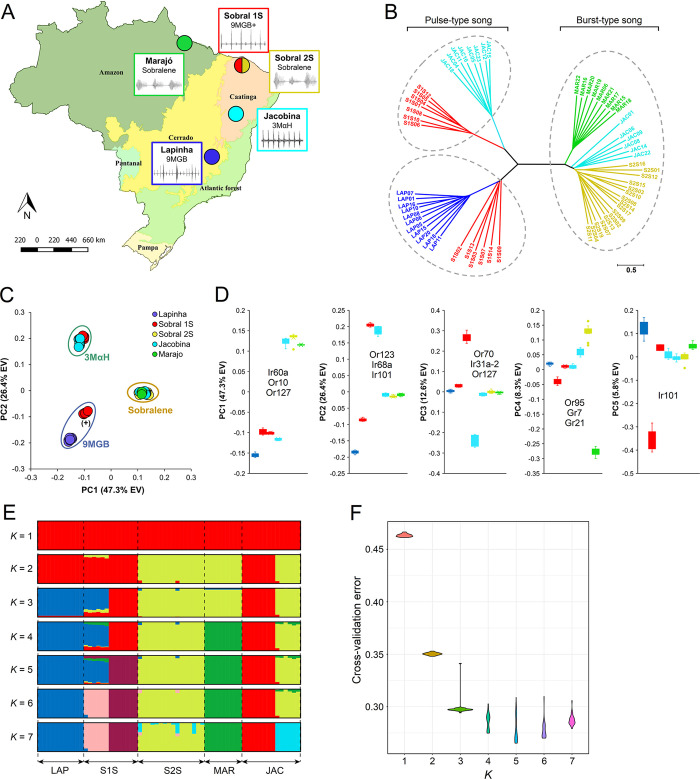
Fig 2. Population structure of 63 L. longipalpis from four sites in Brazil based on SNPs in 245 chemoreceptor genes.
(A) Geographic distribution, sex-aggregation pheromones (9MGB, 3MαH, sobralene) and copulatory songs based on previous studies of L. longipalpis in Brazil (16). (B) Individuals from Sobral (with 2 spots: S2S), Marajo and Lapinha formed discrete clades, while individuals from Sobral (with one spot, S1S) and Jacobina split into two clades each. Unrooted tree estimated using SNPs in the exons of all 100 single-copy orthologs and neighbor-joining method in Tassel v5.2.57. (C) Principal component analysis (PCA) was used to identify loci associated with population structure and conducted using pcadapt (explained variance EV). The first two principal components accounted for 73.6% of the total variation and grouped individuals into four clusters. Putative chemotypes were assigned based on previous studies and PCA clustering patterns. (D) Principal components 1–5 and genes with the highest number of SNPs based on component-wise outlier analysis in pcadapt. (E) Ancestry proportions within individual sand flies for ADMIXTURE models from K = 1 to K = 7 ancestral populations. Each vertical bar represents the proportion of ancestry within a single individual, with colors corresponding to ancestral populations. Data are the average of the major q-matrix clusters derived by CLUMPAK analysis. (F) Violin plot of ADMIXTURE cross-validation error for each of 30 replicates for each K value from 1 to 7.