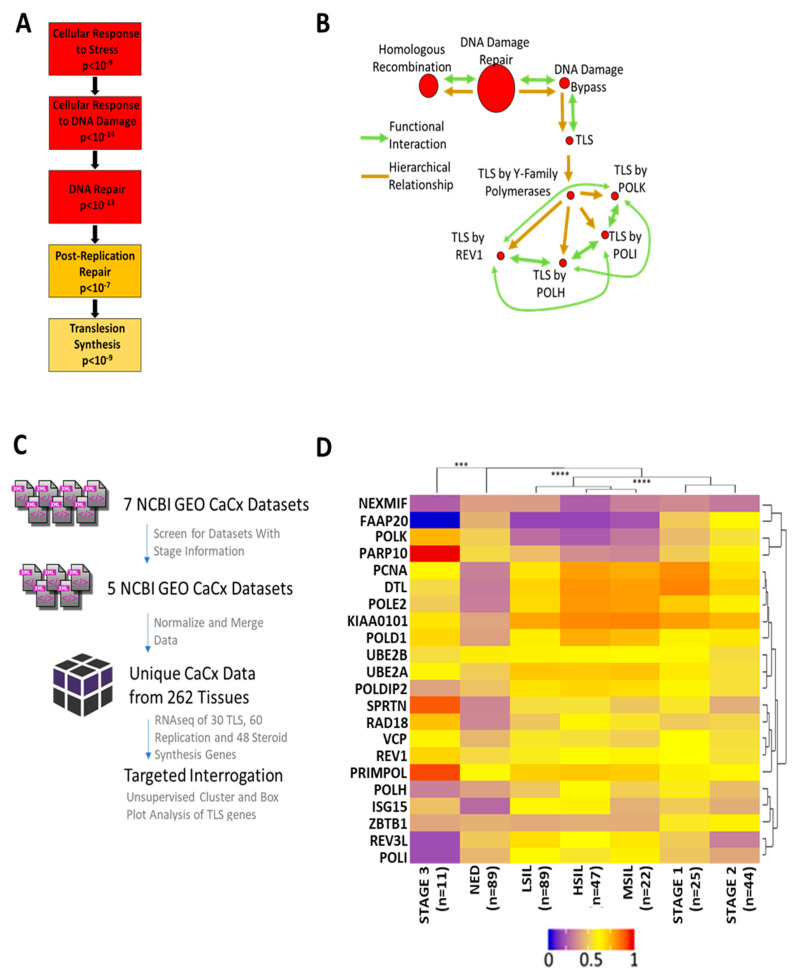
Figure 1.
Computational Analysis of Cervical Cancer Indicates a Dysregulation of TLS Gene Expression. (A) Gene ontology (GO) via GOrilla. Boxes show cellular functions in hierarchical order. Boxes descend from general to specific functions. Darker colors indicate greater enrichment of altered gene expression. (B) GO analysis via GSEA. Lines show hierarchical (gold) and functional (green) relationships. Arrowheads indicate directionality. Pathways are shown as red circles. Larger circles mark broader groupings. (C) Flowchart of data-collection, data-formatting and data-analysis. (D) Heatmap of normalized relative TLS gene expression in CaCx. Colder colors indicate lower relative expression. Warmer colors denote higher relative expression. Unsupervised clustering dendrograms are shown along the x and y axes (*** denotes p < 0.001, **** denotes p < 0.0001; student’s t-test).