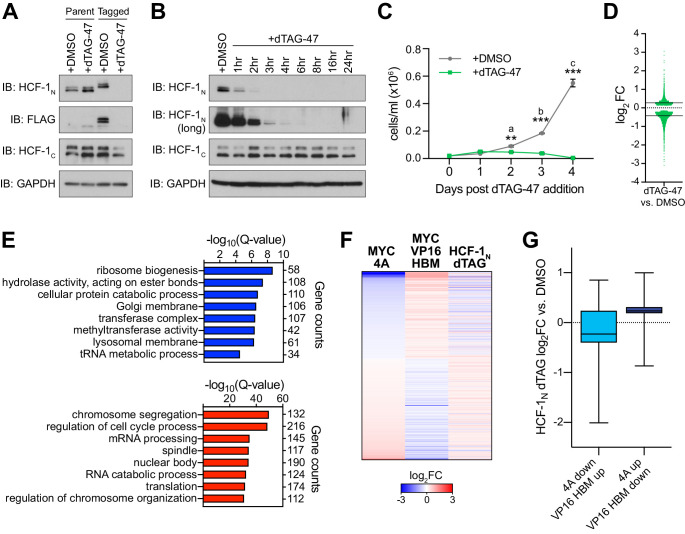
Figure 4. Genes regulated by the MYC−host cell factor (HCF)–1 interaction are impacted by loss of HCF-1.
(A) Western blot, comparing the effects of treating untagged parental cells or FKBPFV-HCF-1N Ramos cells with DMSO or 500 nM dTAG-47 for 24 hr. Blots for HCF-1N, FLAG tag, HCF-1C, and GAPDH are shown. (B) Western blot of lysates from FKBPFV-HCF-1N Ramos cells treated with 500 nM dTAG-47 for varying times, compared to cells treated with DMSO for 24 hr. Shown are short and long exposures of HCF-1N, and HCF-1C, with a GAPDH loading control. (C) Growth curve of FKBPFV-HCF-1N Ramos cells treated with DMSO or 500 nM dTAG-47. Cells were counted every 24 hr for 4 days after plating. Shown are the mean and standard error for three biological replicates. Student’s t-test was used to calculate p-values; a = 0.0029, b = 0.000051, c = 0.000040. (D) Scatterplot showing the distribution of log2FC in RNA-Seq comparing DMSO to 3 hr of 500 nM dTAG-47 treatment (degradation of HCF-1N). Solid lines represent the median log2FC for decreased (−0.425655) and increased (0.270428) transcripts. For clarity, one data point was excluded; this data point is highlighted in Figure 4—source data 2. (E) Gene ontology enrichment analysis of transcripts significantly (false discovery rate [FDR] < 0.05) decreased (top) and increased (bottom) in expression following treatment of FKBPFV-HCF-1N Ramos cells with dTAG-47 for 3 hr. Excluded from this analysis are transcripts that were significantly changed when parental Ramos cells were treated with dTAG-47 for 3 hr. (F) Heatmap showing the log2FC of transcripts with significantly (FDR < 0.05) changed expression, as measured by RNA-Seq, under the indicated conditions. Transcripts are clustered according to the relationship in expression changes between the 4A and VP16 HBM mutants, and ranked by the log2FC for the 4A mutant. Scale of heatmap is limited to [−3,3]. (G) Box-and-whisker plot showing the relationship between transcripts that are anti-correlated between the 4A and VP16 HBM MYC mutants, and significantly changed with the degradation of HCF-1N. Box denotes the 25th to 75th percentile, whiskers extend from minimum to maximum point, and middle line marks the median (4A down/VP16 HBM up: −0.2276; 4A up/VP16 HBM down: 0.2349).