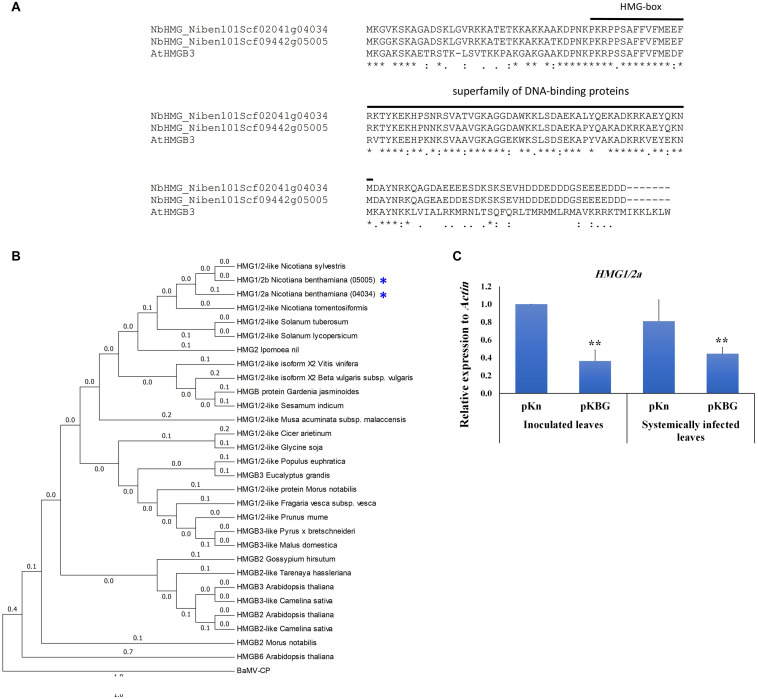
FIGURE 1.
Phylogenetic and expression analyses of Nicotiana benthamiana HMG1/2 genes. (A) Similarities of NbHMG accessions to Arabidopsis thaliana HMGB3. Two NbHMG accessions with high similarity to AtHMGB3 were obtained by blasting the AtHMGB3 coding sequence against the N. benthamiana database (Sol Genomics Networks), i.e., NbHMG_101Scf02041g04034 (NbHMG1/2a) and NbHMG_101Scf09442g05005 (NbHMG1/2b). (B) Phylogenetic analysis of NbHMG1/2 with orthologs from other species. The HMG phylogeny was generated using the Neighbor Joining method in MEGA 6.0 software. Numbers represent relative phylogenetic distance. BaMV capsid protein (CP) was used as an outgroup to root the tree. (C) Relative expression levels of NbHMG1/2a by RTqPCR in N. benthamiana plants infected with BaMV infectious clone (pKBG) compared with plants infiltrated with empty vector (pKn). Inoculated leaves were tested at 3 days post infiltration (dpi), and the systemic leaves were tested at 6 dpi. Actin was used as an internal control. Data represent the mean ± standard deviation from three biological replicates. One-sided student t-tests were performed to determine significant differences. Asterisks indicate significant differences relative to control lines (infiltrated with pKn), with ** representing P < 0.01.