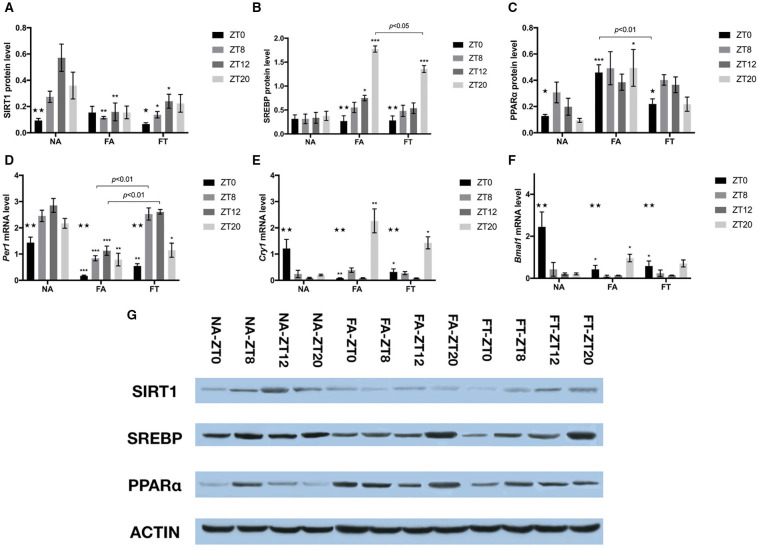
Figure 5.
The effects of feeding regimen on hepatic circadian genes. Hepatic levels of proteins (A–C,G) and mRNAs (D–F) related to circadian rhythm or energy metabolism. Livers were isolated by laparotomy in each group, and the whole cell protein levels of SIRT1, SREBP, and PPARα were measured by Western blotting: (A) SIRT1, Sirtuin1; (B) SREBP, Sterol Regulatory Element Binding Protein; (C) PPARα, Peroxisome Proliferator-activated Receptor α. Relative mRNA expressions were tested using real-time qPCR: (D) Per1 mRNA; (E) Cry1 mRNA; (F) Bmal1 mRNA. Relative amounts of proteins (relative to β-actin) or mRNAs were shown as mean ± SEM. (G) The representative image of immunoblotting analysis of lysates from liver tissue. Data were shown as mean ± SEM (n = 4–5 for each timepoint). Data were compared between groups at same ZT timepoints using one-way ANOVA followed by Bonferroni multiple comparison test. Compared to NA group, *p < 0.05, **p < 0.01, ***p < 0.001. Data were also compared between different ZT timepoints within same feeding regimen using one-way ANOVA, ⋆p < 0.05, ⋆⋆p < 0.01. FA, mice fed a high-fat diet ad libitum; FT, mice fed a time-restricted high-fat diet; NA, mice fed a normal diet ad libitum; ZT, Zeitgeber time.