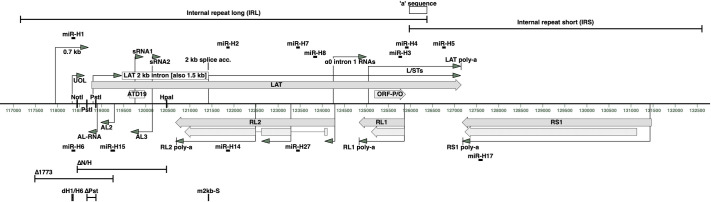
Fig 1. Schematic of the HSV-1 LAT locus RNAs.
The LAT locus is duplicated in the TRL and IRL inverted repeats of the HSV-1 genome, here showing IRL next to IRS that together span 117940:127160 in HSV-1 strain 17syn+ genome (GenBank accession JN555585.1). The latency-associated transcript promoter LAP1 is followed by an enhancer element LAP2. The primary 8.3 kb LAT transcript has two exons and a 2.0 kb intron. These, together with multiple other RNAs (0.7 kb, UOL, sRNA1 and 2, α0 intron 1 RNAs 1–4, and L/STs giving rise to the ORF-P and ORF-O proteins) and miRNAs (miR-H1, H2, H7, H8, H3, H4, and H5) transcribed from within or partially overlapping the LAT locus are known as LATs. The RL2 and RL1 genes (yielding the ICP0 and ICP34.5 proteins) are located anti-sense to LAT. In addition, many RNAs (AL-RNA, AL2, and AL3) and miRNAs (miR-H6, H15, H14, and H27) have been reported to map anti-sense to the LAT locus. The RS1 gene (ICP4 protein) and miR-H17 are on the opposite strand in IRS. The LAT, RS1, RL1, and RL2 polyadenylation sites are indicated. The LAT probe ATD19 binding site is indicated. Changes in five LAT mutant viruses used in the study are marked. The 17ΔN/H deletion spans NotI-HpaI (118447:120472), containing LAP1, exon 1, and part of the 2 kb intron. The Δ1773 deletion removes sequences prior to LAP1 and part of exon 1 (117486:119258). 17dmiR-H1/H6 (dH1/H6) has a 25 nt deletion removing both miR-H1-5p and miR-H6-3p (118328:118352). m2kb-S contains three mutations at the LAT 2 kb intron splice acceptor site (at 121420, 121421, and 121424 nt). The KOSΔPstLAT deletion spans PstI-PstI (118558:118760).