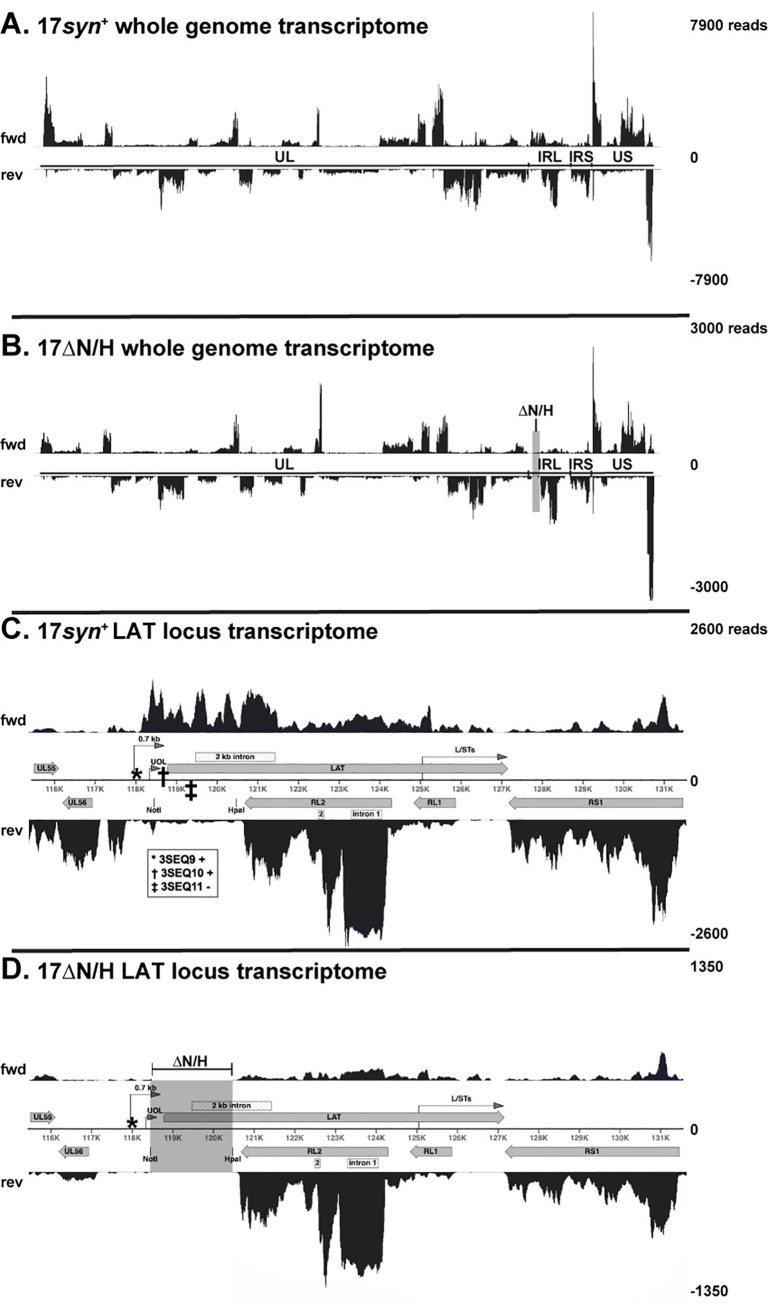
Fig 5. Viral transcriptome and LAT locus transcription in skin xenografts infected with 17syn+ and 17ΔN/H.
Total RNA was extracted from 17syn+- or 17ΔN/H-infected (2,500 PFU) skin xenografts at 11 DPI and pooled for RNA-seq. Reads were mapped to a reference genome consisting of UL-IRL-IRS-US, with the inverted repeats TRL and TRS omitted. The indicated read scales were determined by the maximum peak of reads. Histograms of strand-specific (fwd or rev) transcription coverage across (A) 17syn+ and (B) 17ΔN/H genomes are shown. The ΔN/H deletion is highlighted by a grey box. A close up of LAT locus transcription is shown for (C) 17syn+ and (D) 17ΔN/H. The area shown includes two preceding UL genes (UL55 and UL56), IRL genes (LAT, RL1, and RL2), and an IRS gene (RS1). Additional region hits for polyadenylated transcript ends found using 3SEQ are marked as * 3SEQ9 (fwd), † 3SEQ10 (fwd), and ‡ 3SEQ11 (rev) (which are not annotated in GenBank accession JN555585.1). Two of these, 3SEQ10 and 3SEQ11, are within the region deleted in the 17ΔN/H. Additional transcripts shown include the L/STs that end at the LAT polyadenylation site, as well as 0.7 kb LAT and UOL for which the polyadenylation sites could be the source of the 3SEQ10 hit.