Abstract
In 2016, the WHO included haemoglobin values within normal ranges as a diagnostic criterion for Polycythaemia Vera (PV). Since then, concerns have arisen that a large number of patients are undergoing unnecessary screening for PV. To address this issue, we estimated the prevalence of JAK2 p.V617F in individuals with elevated haemoglobin or haematocrit and developed and validated a screening algorithm for PV. A total of 15,366 blood counts performed in seven non-consecutive days were reviewed, of which 1001 were selected for subsequent JAK2 p.V617F mutation screening. Eight (0.8%) new JAK2 p.V617F-mutated cases were detected. From ROC curves, a two-step algorithm was developed based on the optimal cut-off for the detection of the JAK2 p.V617F mutation. The algorithm was prospectively validated in an independent cohort of 15,298 blood counts. A total of 1595 (10.4%) cases met the criterion for haemoglobin or haematocrit, of whom 581 passed to step 2 (3.8% of the total). The JAK2 p.V617F mutation was detected in 7 of the 501 patients tested, which accounts for 0.04% of the total cohort and 0.4% of patients with erythrocytosis. In conclusion, this data show that our two-step algorithm improves the selection of candidates for JAK2 p.V617F testing.
Subject terms: Molecular medicine, Oncology, Cancer, Cancer genetics, Cancer screening, Haematological cancer, Biomarkers, Diagnostic markers, Diagnostic markers, Medical research, Translational research
Introduction
The presence of the JAK2 p.V617F mutation one of the major criterion for the diagnosis of Polycythaemia Vera (PV), although it is found in other myeloproliferative neoplasms1. Another major diagnostic criterion is elevated haemoglobin (Hb) concentration (> 16.5 g/dL in men; > 16 g/dL in women) or elevated haematocrit (Htc) (> 49% in men; > 48% in women) or increased red blood cell mass1. The thresholds for haemoglobin and haematocrit were modified in the 2016 World Health Organization (WHO) update. These thresholds were modified after it was observed that 40% of cases of PV were misdiagnosed when the 2008 WHO Hb thresholds were used and red cell mass or bone marrow biopsy was not performed2. However, some studies suggest that the 2016 WHO haemoglobin thresholds could lead to unnecessary testing. Thus, with these thresholds, the number of men and women tested has been calculated to have increased 12-fold and threefold, respectively, which also has a high psychosocial burden3,4. In order to provide a solution, some authors recommend that higher haemoglobin cut-off values are used, or that the diagnostic algorithm is based on the combined presence of thrombocytosis, neutrophilia or previous thrombosis3,5. Nonetheless, even when these diagnostic approaches are applied, a high proportion of cases will be still missed, with a high rate of false negatives (14–64%)6.
Several recent reports highlight the relevance of detecting JAK2 p.V617F clonal haematopoiesis, since it is strongly associated with an increased cardiovascular risk7,8. In large cohort studies, the prevalence of JAK2 p.V617F in the general population has been estimated to range between 0.1 and 3.1%. This variability can be explained by differences in the sensitivity of the tests used for the detection of the JAK2 mutation9–12. Thus, the prevalence of JAK2 p.V617F with a ≥ 1% allele burden ranges between 0.1 and 0.5%10–13. However, no data is currently available on the prevalence of JAK2 p.V617F in individuals with increased Hb or Htc, and an effective diagnostic approach has not yet been established.
To address this problem, we developed and validated a sequential two-step-algorithm screening test based on the prevalence of JAK2 p.V617F in individuals with elevated levels of haemoglobin or haematocrit according to WHO 2016 criteria.
Methods
Study design and sample collection
This study was approved by the Institutional Review Board of Hospital Ramón y Cajal, Spain. Informed consent was obtained from JAK2 p.V617F-positive patients in accordance with the Declaration of Helsinki. All JAK2 p.V617F-positive patients were contacted. Standard diagnostic work-up was performed in accordance with relevant guidelines and regulations. The need for informed consent from JAK2 p.V617F-negative individuals was waived by the approving ethics committee. None of the authors had access to the identity of JAK2 p.V617F-negative individuals during data analysis.
The study was divided into two phases: a first Phase, which involved the development of an algorithm based on the prevalence of JAK2 p.V617F in individuals with elevated Hb or Htc; and a second Phase where the algorithm was validated in an independent cohort. Blood samples were collected in EDTA tubes and analysed in a CELL-DYN Sapphire analyser (Abbott). The following parameters were measured in all samples: haemoglobin (Hb), haematocrit (Htc), leukocytes, neutrophils, platelets, mean corpuscular volume (MCV), mean corpuscular haemoglobin (MCH), and red cell distribution width (RDW).
Phase 1
A total of 15,366 blood samples were prospectively analysed in seven non-consecutive days. The samples that met the 2016 WHO criteria for Hb or Htc (males with Hb > 16.5 g/dL or Htc > 49%, and females with Hb > 16 g/dL or Htc > 48%) were selected for subsequent JAK2 p.V617F mutation screening. ROC curves were used to estimate the sensitivity and specificity of leukocyte count, neutrophil count, platelet count MCV, RDW, MCH, and MCHC for detecting the JAK2 p.V617F mutation. The variables with the best diagnostic accuracy were included in a sequential two-step-algorithm.
Phase 2
The two-step-algorithm was validated in blood samples from an independent cohort of 15,298 individuals. Only the samples that met the criteria of the two-step-algorithm were tested for JAK2 p.V617F.
JAK2 p.V617F was also tested in 300 unselected samples obtained in an outpatient setting in primary care centres to determine the prevalence of JAK2 p.V617F clonal haematopoiesis in our environment.
JAK2 p.V617F mutation screening
Genomic DNA was extracted from peripheral blood samples by the Chemagic MSM-I automated magnetic bead method. JAK2 p.V617F screening was performed using a novel, allele-specific multiplex PCR assay. In this assay, we achieved the specific amplification of a 135-bp amplicon only in carriers of the c.1849G > T PV mutation by using primers 5′-GCATTTGGTTTTAAATTATGGAGTATGCT-3′ and 5′-ACACCTAGCTGTGATCCTGAAACTGA-3′. We simultaneously amplified a 291-bp genomic fragment encompassing JAK2 exon 16. This was an internal control to rule out false negative results due to PCR failure by using primers 5′- GGCTTGAACATACTAAATGCTCCAGTA-3′ and 5′-AAGGAAAATTAACAACATGCCCTTTAC-3′. PCR was carried out using the following process: a cycle of denaturation at 94 °C for 2 min; five touchdown cycles of denaturation at 94 °C for 30 s; annealing for 30 s at 65 °C for the first cycle; a 1 °C reduction per cycle; and extension at 72 °C for 30 s; 25 cycles of denaturation at 94 °C for 30 s; annealing at 60 °C for 30 s; extension at 72 °C for 30 s; and a final extension step of 72 °C for seven minutes. The multiplex reaction with both primer pairs took place at a final concentration of 1.5 mM MgCl2 with Fast Start Taq DNA polymerase (Roche). PCR products were resolved in a TapeStation 2200 microfluidics electrophoresis device (Agilent).
In our assay, a threshold of mutational burden was established for the reliable detection of the p.V617F allele. To such purpose, we analysed serial dilutions of a control sample with a known p.V617F allele burden. It was confirmed that our assay consistently detected samples with mutational burdens ≥ 1% as positive for JAK2 p.V617F.
All positive results from the multiplex assay were confirmed by quantitative real-time PCR in the Laboratory of Molecular Biology of Hospital 12 de Octubre. As previously published, samples with an allele burden > 0.71% were considered positive14.
Statistical analyses
Univariate analysis was performed to assess the relationship between the variables analysed and the result of the JAK2 test. Since all parameters, except for cell-count values (leukocytes, neutrophils and platelets), followed a normal distribution, differences were assessed using a parametric test. Student t-test was used for quantitative variables, whereas differences between categorical variables were assessed by the Chi-squared test. In relation to cell-count variables, a non-parametric test (Mann–Whitney U) was used. All p values < 0.05 were considered significant. All data were analysed using SPSS 20.0 (IBM corp.) statistical software.
Results
Phase 1
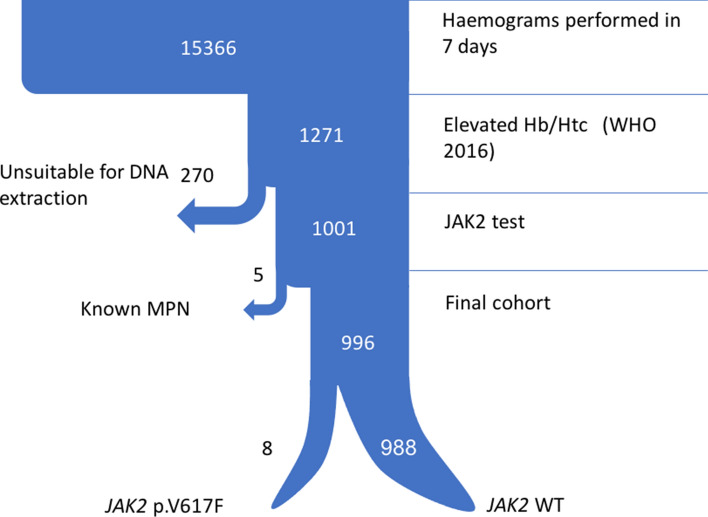
A total of 15,366 blood samples were analysed, of which 1271 (8.3%) showed elevated Hb or Htc levels according to WHO 2016 criteria, and were selected for JAK2 testing. As many as 1001 of these samples were tested for JAK2 p.V617F, of which 13 were positive (1.3%). The clinical records of the 13 JAK2-positive cases were reviewed, of which 5 were excluded because they corresponded to patients with a known myeloproliferative neoplasm. Therefore, the final prevalence of the JAK2 p.V617F mutation in the 996 patients who met WHO Hb or Htc criteria was 0.8% (8/996). A sample flowchart is shown in Fig. 1.
Figure 1.
Step 1 sample flowchart. This figure contains a diagram of the sample selection process. WHO World Health Organization, MPN myeloproliferative neoplasm, WT wild type.
The mean and median values for all the parameters studied in each group are shown in Table 1. The exclusion of 275 samples (270 were unsuitable for DNA extraction and 5 had a previous diagnosis of myeloproliferative neoplasm) had no impact on the characteristics of the groups (Table 1).
Table 1.
Characteristics of study groups.
| All (N = 15,366) | WHO (N = 1271) | JAK2 (N = 996) | |
|---|---|---|---|
| Age | 60.55 (0–104) | 57.83 (14–100) | 58.17 (14–100) |
| Hb (g/dl) | 13.43 (4.1–21) | 16.5 (13.4–21) | 16.5 (13.4–21) |
| Haematocrit (%) | 40.76 (12.3–65) | 50.63 (36.4–65) | 50.52 (36.4–65) |
| WBC (× 109/L) | 7.79 (0–254) | 8.36 (2.2–43.5) | 8.25 (2.2–27.1) |
| Neutrophils (× 109/L) | 4.63 (0–160) | 4.75 (0.9–40.9) | 4.63 (0.9–22.4) |
| Platelets (× 109/L) | 233.15 (2–1096) | 223.35 (37–734) | 224.41 (69–734) |
| MCV (fL) | 91.59 (52–139) | 92.73 (69–119) | 92.56 (69–112) |
| RDW | 13.43 (10–37) | 12.98 (11–22) | 12.92 (11–22) |
| MCH | 30.21 (15–45) | 30.26 (22–47) | 30.26 (22–47) |
| MCHC | 32.98 (25–50) | 32.65 (25–46) | 32.70 (25–46) |
WHO individuals meeting World Health Organization 2016 Hb or Htc criterion, JAK2 patients in which JAK2 p.V617F was studied, Hb haemoglobin, WBC white blood cells, MCV mean corpuscular volume, RDW red cell distribution width, MCH mean corpuscular haemoglobin, MCHC mean corpuscular haemoglobin concentration.
Algorithm development process
In order to identify the markers that could be useful to identify patients with the JAK2 p.V617F mutation, a univariate analysis was performed according to the mutational status of JAK2 (Table 2).
Table 2.
Univariate analysis according to JAK2 mutational status.
| JAK2 p.V617F (n = 8) | JAK2 WT (n = 988) | p-value | |
|---|---|---|---|
| Sex | |||
| Female | 1 (16.7%) | 203 (20.5%) | 0.816 |
| Male | 5 (83.3%) | 787 (79.5%) | |
| Age | 69.5 (18.3) | 58 (9.6) | 0.079 |
| Haemoglobin | 16.5 (1.2) | 16.5 (0.8) | 0.925 |
| Haematocrit | 51.3 (3.2) | 50.5 (2.1) | 0.317 |
| WBCb median (p.25;p75) | 10.6 (8.2;11.6) | 7.8 (6.6;9.2) | 0.022 |
| Neutrophilsb median (p.25;p75) | 6.4 (4.7;7.8) | 4.2 (3.3;5.4) | 0.015 |
| Plateletsb median (p.25;p75) | 324 (272.5;489) | 217 (183;253) | < 0.001 |
| MCV | 89.1 (4.7) | 92.6 (4.6) | 0.033 |
| RDW | 15.3 (3.1) | 12.9 (1.2) | < 0.001 |
| MCH | 28.6 (1.7) | 30.3 (1.9) | 0.015 |
| MCHC | 32.1 (1.1) | 32.7 (1.4) | 0.247 |
WBC white blood cells, MCV mean corpuscular volume, RDW red cell distribution width, MCH mean corpuscular haemoglobin, MCHC mean corpuscular haemoglobin concentration.
aT-Student or Chi-square as appropriate.
bNon parametric U-Mann–Whitney.
We found that patients positive for the JAK2 p.V617F mutation had statistically significant (p < 0.05) higher levels of leukocytes, neutrophils, platelets and RDW, and lower levels of MCV and MCH, as compared to JAK2 p.V617F-negative patients (Table 2). There was a tendency in patients with JAK2 p.V617F to be older, although differences did not reach statistical significance (p = 0.079).
For the variables with statistically significant differences, we calculated the area under the ROC curve (AUC) and the optimal cut-off points that maximize the sensitivity and specificity of the test (Youden index) (Table 3).
Table 3.
Area under de ROC curve and optimal cut-off point.
| AUC | Optimal cut-off | Sensibility | Specificity | |
|---|---|---|---|---|
| WBC | 0.72 | 9.92 | 0.63 | 0.81 |
| Neutrophils | 0.79 | 5.98 | 0.75 | 0.83 |
| Platelets | 0.86 | 248.5 | 1.00 | 0.72 |
| MCV | 0.50 | 79.55 | 1.00 | 0.01 |
| RDW | 0.80 | 13.05 | 1.00 | 0.60 |
| MCH | 0.51 | 25.40 | 1.00 | 0.02 |
AUC area under the curve, WBC white blood cells, MCV mean corpuscular volume, RDW red cell distribution width, MCH mean corpuscular haemoglobin.
Of interest, neutrophils, platelets and RDW showed a high AUC (> 0.75). With a neutrophil cut-off of 5.98 × 109/L, sensitivity and specificity were 75% and 83%, respectively. A platelet cut-off of 248.5 × 109/L and a RDW cut-off of 13.05 showed 100% sensitivity. From this data, we developed a screening algorithm based on thresholds for platelets and neutrophils, given their statistical, clinical, and biological significance (Fig. 2).
Figure 2.
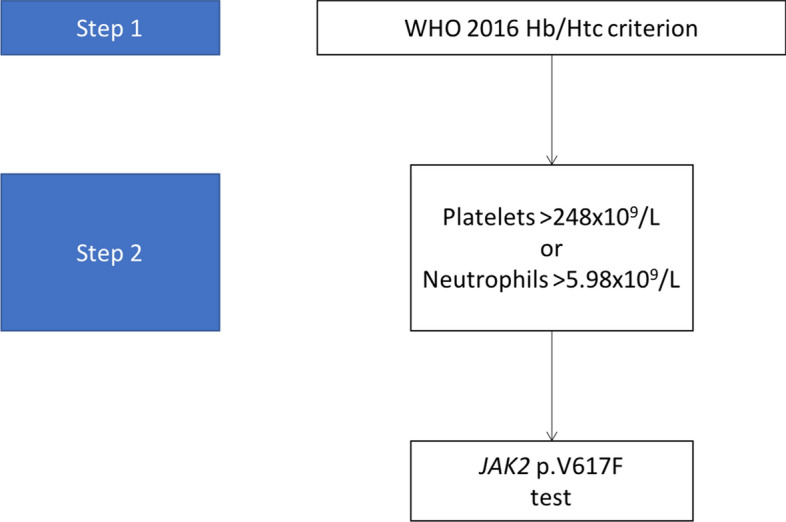
The “two-step” algorithm. This figure shows the algorithm based on data obtained in Step 1.
Phase 2
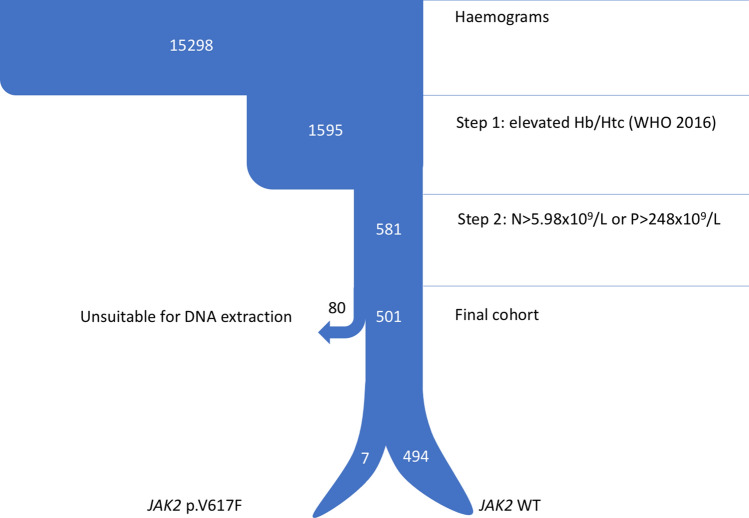
The proposed algorithm was prospectively validated in an independent cohort composed of 15,298 individuals. A total of 1595 (10.4%) patients exhibited elevated Hb or Htc levels according to the 2016 WHO criteria, of whom 581 passed to step 2 (3.8% of the total) and were selected for JAK2 p.V617F testing. Of the latter, 80 were not suitable for DNA extraction. The JAK2 p.V617F mutation was detected in seven of the 501 patients tested, which accounts for 0.04% of the total cohort, 0.4% of patients with elevated Hb or Htc levels, and 1.2% of the patients who fulfilled the two-step algorithm. None of these patients had a previous diagnosis of myeloproliferative neoplasm. A flowchart of samples is shown in Fig. 3.
Figure 3.
Step 2 sample flowchart. This figure contains a diagram of the samples selected in the different steps of the algorithm. N neutrophils, P platelets, WHO World Health Organization, WT wild type.
A detailed description of the characteristics of the patients who passed Steps 1 and 2 is provided in Table S1 of online Supplementary material.
To confirm the prevalence of JAK2 p.V617F mutation in our environment, samples from 300 unselected individuals were assayed for p.V617F, with 1 positive result that corresponded to a patient with a previous diagnosis of myeloproliferative neoplasm. Therefore, the prevalence rate was at least < 1/300.
Discussion
The current WHO diagnostic criteria for Polycythaemia Vera include Hb and Htc values below the threshold established in the definition of erythrocytosis. This raises the question of whether all these patients should be tested for the JAK2 p.V617F mutation. The results of this study show that an average of 1500 blood samples would be eligible every week for JAK2 p.V617F testing, with a rate of positive results as low as 0.4%. This protocol involves an excessive workload and our results highlight that other criteria are needed for the selection of samples eligible for JAK2 p.V617F screening.
In the present work, we have designed a two-step algorithm that would reduce JAK2 p.V617F testing to 500 samples per week. The proposed algorithm is based on blood count parameters that would increase pre-test specificity. Given that JAK2 p.V617F-positive diseases cause myeloproliferation and PV is associated with iron deficiency secondary to erythropoiesis, we selected parameters that identified either of the two phenomena. Based on these parameters and their optimal cut-off points, we built a two-step algorithm. The first step involves the identification of males with Hb > 16.5 g/dl or htc > 49% or females with Hb > 16 g/dl or htc > 48%. The second step consists of identifying among the selected patients those who had either neutrophils > 5.98 × 109/L or platelets > 248.5 × 109/L. However, to facilitate the use of the algorithm, we rounded thresholds to 6 × 109/L for neutrophils and 250 × 109/L for platelets (Fig. 4).
Figure 4.
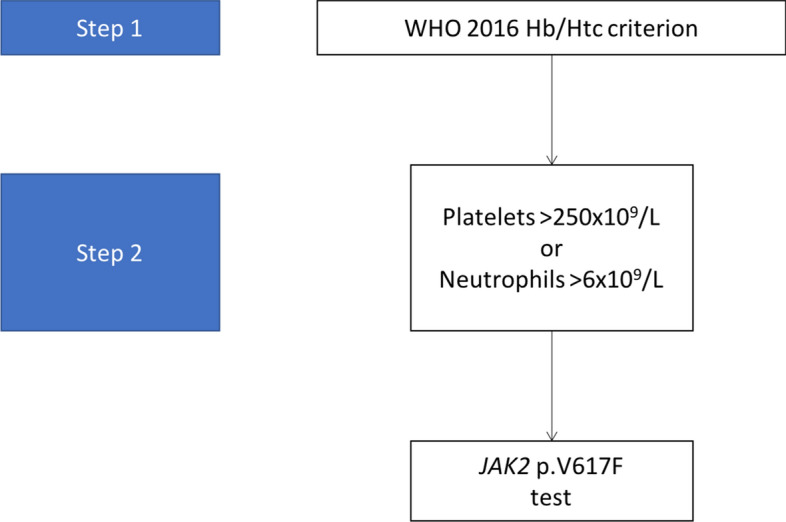
Simplified two-step algorithm. This figure shows a simplified version with rounded threshold values.
Finally, the “two-step” algorithm was prospectively validated in an independent cohort of patients. However, the rate of positivity of JAK2 p.V617F in this cohort was only 1.2%, a value too low to be used in routine practice, unless massive JAK2 p.V617F screening platforms are developed.
Others approaches have been used in retrospective studies. Rumi et al. suggested testing for JAK2 p.V617F in the presence of elevated Hb or Htc levels in parallel to increased neutrophils or platelets. Alternatively, if elevated Hb levels are considered alone, the threshold for screening can be increased to 17 g/dL in men6. However, our data showed that increasing the Hb threshold alone would not be effective, since Hb levels were below 17 g/dL in most of our JAK2 p.V617F-positive patients (73.4%). The Canadian proposal discourages testing when other signs of myeloproliferation are present, such as platelets > 440 × 109/L or neutrophils > 7 × 109/L4. Again, if we apply that approach to our series, only 30% of our cases would meet the criteria, which indicates that the approach is suboptimal.
It is important to note that a third of patients found to be JAK2 p.V617F-positive in the phase of prospective validation of Step 2 had suffered a cardiovascular event. Moreover, in a recent study, the presence of clonal haematopoiesis of undetermined significance in peripheral blood cells was associated with an increased risk of coronary heart disease, with the highest risk corresponding to the JAK2 p.V617F mutation9. These findings support JAK2 p.V617F screening in the general population and emphasize the need for an improved approach to the identification of potential candidates for JAK2 p.V617F screening.
A limitation of the present study is that the prevalence of the JAK2 p.V617F mutation was remarkably lower, as compared to previous studies. Recent data has shown that the prevalence of the JAK2 p.V617F mutation in the general population can be as high as 3.1% when a very sensitive test is used (> 0.009%)10. Nevertheless, the prevalence of JAK2 p.V617F with ≥ 1% of allele burden decreased to 0.5%, which is consistent with the results published by other groups. Our results indicate a prevalence of 0.4% of the JAK2 V617F mutation in individuals with elevated Hb or Htc levels.
In conclusion, our data show that WHO thresholds for Hb and Htc for the diagnosis of PV should not be considered alone for JAK2 p.V617F screening. The two-step algorithm proposed in this study improves the selection of candidates for JAK2 p.V617F testing.
Supplementary Information
Acknowledgements
The authors thank the staff of the haematology laboratory of Ramón y Cajal Hospital, with special thanks to JL Moncada, JM del Rey, and J Zamora, who performed the statistical analysis.
Author contributions
M.P.V., V.G.G. and J.M.L. designed the study. M.P.V., A.S.M. and C.N.T. selected the samples. G.M.M., F.J.C. and R.S. were responsible for laboratory work and participated in data analysis and interpretation. M.P.V., V.G.G., A.A.L., J.V., J.L.J. and J.M.L. participated in data analysis and interpretation. M.P.V. and A.A.L. drafted the manuscript. M.P.V. designed the figures. All authors contributed to the final approved version of this manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Miguel Piris-Villaespesa, Email: mpirisv@gmail.com.
Valentin Garcia-Gutierrez, Email: jvalentingg@gmail.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-020-80459-y.
References
- 1.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez-Larran A, Ancochea A, Angona A, Pedro C, Garcia-Pallarols F, Martinez-Aviles L, et al. Red cell mass measurement in patients with clinically suspected diagnosis of polycythemia vera or essential thrombocythemia. Haematologica. 2012;97(11):1704–1707. doi: 10.3324/haematol.2012.067348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Busque L, Porwit A, Day R, Olney HJ, Leber B, Éthier V, et al. Laboratory investigation of myeloproliferative neoplasms (MPNs): Recommendations of the Canadian MPN group. Am. J. Clin. Pathol. 2016;146(4):408–422. doi: 10.1093/ajcp/aqw131. [DOI] [PubMed] [Google Scholar]
- 4.Sandes AF, Gonçalves MV, de Chauffaille M. Frequency of polycythemia in individuals with normal complete blood cell counts according to the new 2016 WHO classification of myeloid neoplasms. Int. J. Lab. Hematol. 2017;39(5):528–531. doi: 10.1111/ijlh.12686. [DOI] [PubMed] [Google Scholar]
- 5.Rumi E, Cazzola M. Diagnosis, risk stratification, and response evaluation in classical myeloproliferative neoplasms. Blood. 2017;129(6):680–692. doi: 10.1182/blood-2016-10-695957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbui T, Thiele J, Gisslinger H, Carobbio A, Vannucchi AM, Tefferi A. Diagnostic impact of the 2016 revised who criteria for polycythemia vera. Am. J. Hematol. 2017;92(5):417–419. doi: 10.1002/ajh.24684. [DOI] [PubMed] [Google Scholar]
- 7.Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014;371(26):2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N. Engl. J. Med. 2017;377(2):111–121. doi: 10.1056/NEJMoa1701719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordua S, Kjaer L, Skov V, Pallisgaard N, Hasselbalch HC, Ellervik C. Prevalence and phenotypes of JAK2 V617F and calreticulin mutations in a Danish general population. Blood. 2019;134(5):469–479. doi: 10.1182/blood.2019001113. [DOI] [PubMed] [Google Scholar]
- 10.Nielsen C, Birgens HS, Nordestgaard BG, Bojesen SE. Diagnostic value of JAK2 V617F somatic mutation for myeloproliferative cancer in 49,488 individuals from the general population. Br. J. Haematol. 2013;160(1):70–79. doi: 10.1111/bjh.12099. [DOI] [PubMed] [Google Scholar]
- 11.McKerrell T, Park N, Moreno T, Grove CS, Ponstingl H, Stephens J, et al. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep. 2015;10(8):1239–1245. doi: 10.1016/j.celrep.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu X, Zhang Q, Luo J, Xing S, Li Q, Krantz SB, et al. JAK2V617F: Prevalence in a large Chinese hospital population. Blood. 2007;109(1):339–342. doi: 10.1182/blood-2006-03-009472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langabeer SE. An increase in diagnostic JAK2 V617F mutation testing: Is masked polycythaemia vera the explanantion? Eur. J. Intern. Med. 2018;52:e37–e38. doi: 10.1016/j.ejim.2018.03.017. [DOI] [PubMed] [Google Scholar]
- 14.Rapado I, Albizua E, Ayala R, Hernández JA, Garcia-Alonso L, Grande S, et al. Validity test study of JAK2 V617F and allele burden quantification in the diagnosis of myeloproliferative diseases. Ann. Hematol. 2008;87(9):741–749. doi: 10.1007/s00277-008-0512-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.