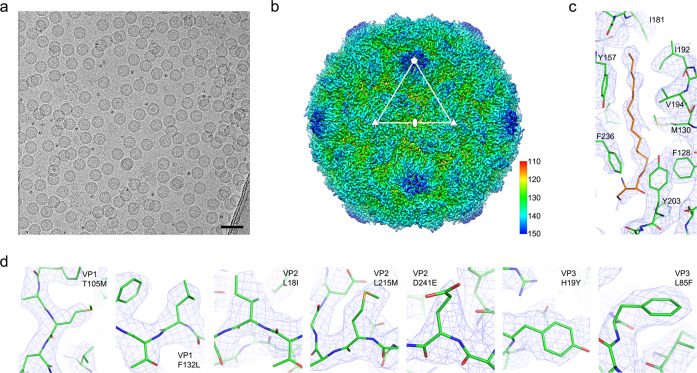
Fig. 8. Cryo-EM structure analysis of the stabilised PV3 SC8 VLP.
a Cryo-electron micrograph of PV3 SC8 VLPs embedded in vitreous ice. Scale bar 50 nm. b Cryo-EM map of the PV3 SC8 reconstruction coloured by radius from red to blue according to the scale bar shown. Representative 5-fold, 3-fold and 2-fold axes are indicated. The outer diameter of the particle is ~300 Å. c Electron potential map for the PV3 SC8 VP1 pocket showing the bound sphingosine-like molecule (orange) and surrounding residues of VP1 (green); key residues interacting with the modelled sphingosine are labelled. Electron potential is contoured at 1.0 σ and rendered at a radius of 2 Å around atoms. d Individual panels showing the cryo-EM density for the stabilising mutations introduced into the PV3 SC8 VLP. Residue numbering is equivalent to the mature capsid proteins VP1, VP2 and VP3. VP4 T67A was disordered. Electron potential is contoured at 1.0 σ and rendered at a radius of 2 Å around atoms.