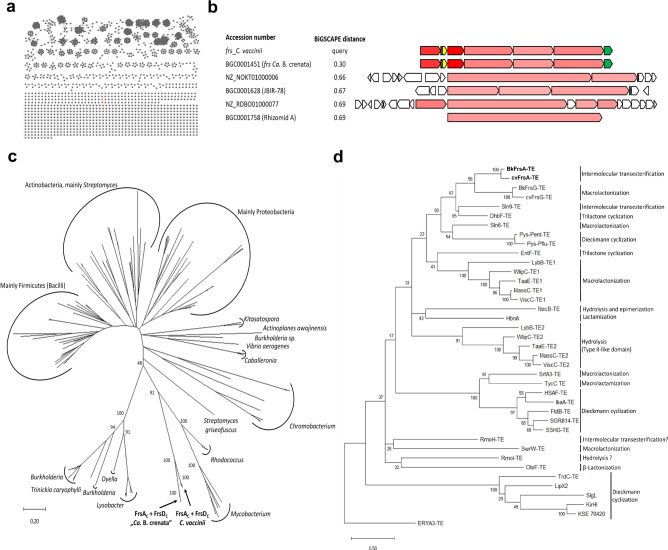
Fig. 5. Bioinformatic analyses of the frs BGC and selected domains.
a BiG-SCAPE analysis network of frs and 2716 NRPS BGCs, obtained from a global BiG-SLiCE query against all non-fragmented NRPS from the BiG-FAM database (distance of d < 1400) at distance cutoff 0.5. The frs BGCs from C. vaccinii and “Ca. B. crenata” (red) do not cluster with any other BGC. b Alignment of frs and their 4 closest BGCs from the BiG-SCAPE analysis in Fig. 5a. Color intensity denotes the degree of homology to the respective protein of the frs BGC. Red: frsD (NRPS), blue: frsB (MbtH-like protein), yellow: frsC (dehydrogenase), green: frsH (hydroxylase). None of the listed BGCs contains homologues to frsC or frsH. Alignment adapted from Corason45. c Phylogenetic tree of Cstarter domains. Taxonomy of clades is indicated. The scale bar represents 20 substitutions per 100 amino acids. d Phylogenetic tree of NRPS TE domains. The observed mode of peptide release is indicated. The scale bar represents 50 substitutions per 100 amino acids.