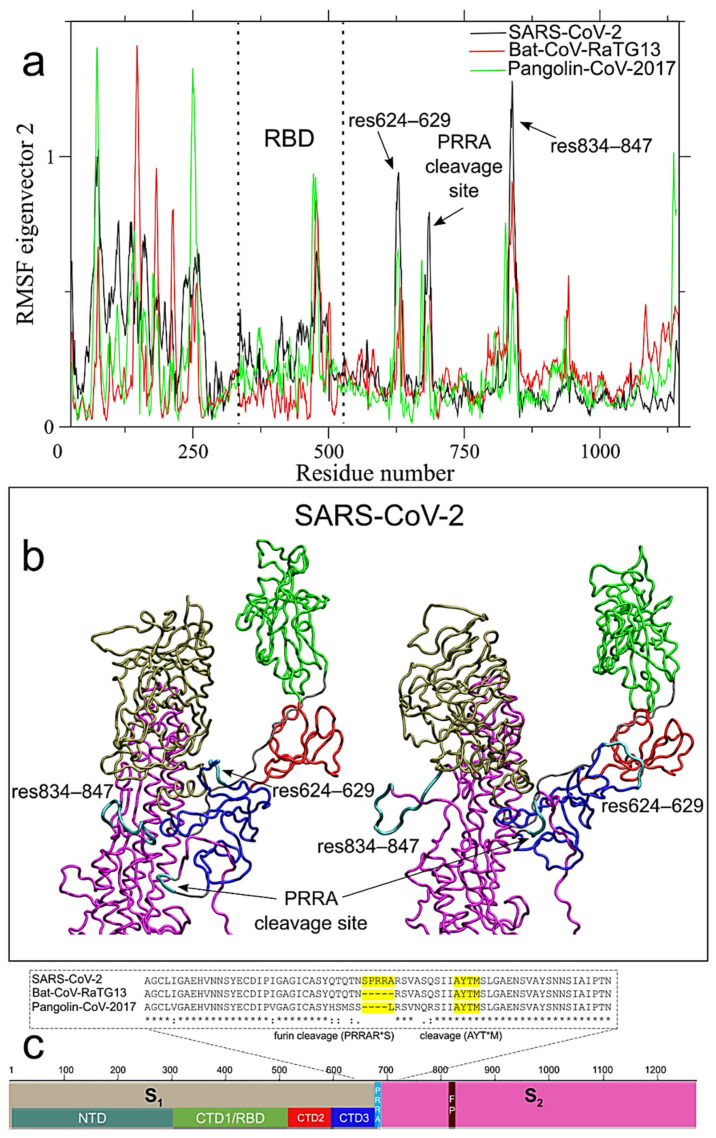
Figure 3.
Long-range correlated motions along ED eigenvector 2. (a) Per-residue Root Mean Square Fluctuations (RMSF) in nm of the S protein filtered trajectory along ED eigenvectors 2 for SARS-CoV-2, Bat-CoV-RaTG13 and Pangolin-CoV-2017 are reported in black, red and green colors, respectively. Residues corresponding to peak of fluctuations in SARS-CoV-2 are reported. (b) Projection of the S protein MD trajectory along ED eigenvector 2 for the SARS-CoV-2 system. Residues 624–629, the newly acquired PRRA cleavage site and res834–847, corresponding to peak of fluctuations shown in panel (a), are highlighted in cyan color. (c) Local alignment in the PRRA cleavage site. In the alignment subsets, conserved residues are marked by “*”; “.” indicates that one strain is different from SARS-CoV-2 at the marked position; “:” indicates both other strains differ from SARS-CoV-2.