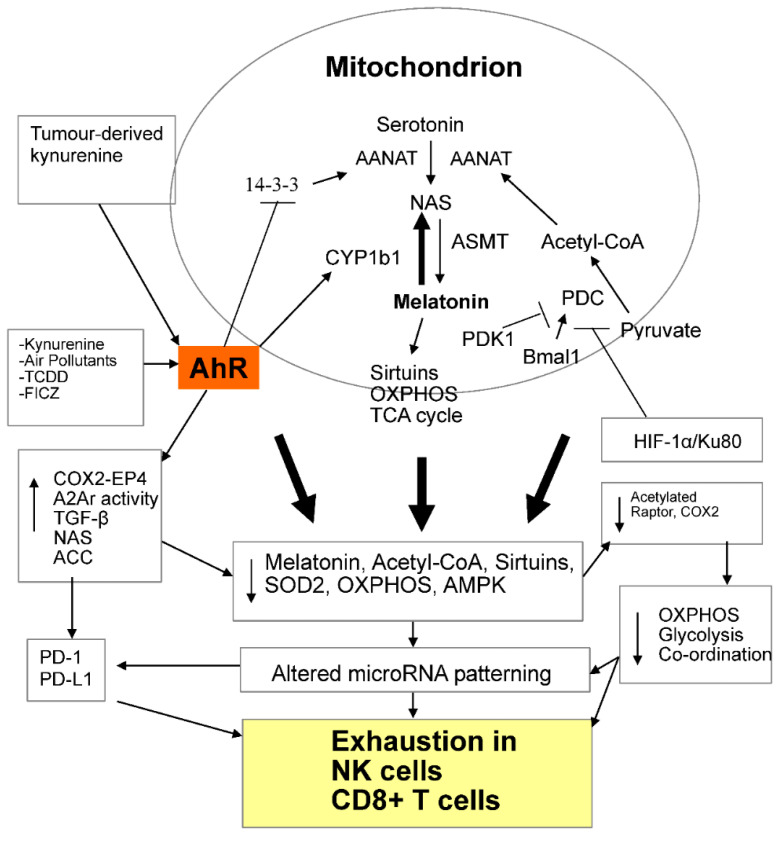
Figure 1.
Shows how AhR ligands, such as air pollutants, FICZ, TCDD and kynurenine, including tumour derived, activate the AhR to modulate mitochondrial metabolism, partly via the regulation of the melatonergic pathway. The melatonergic pathway is shown in the mitochondrion, but is also present in the cytoplasm. AhR activation drives a number of processes and factors underpinning exhaustion and tumour survival, including the COX2/PGE2/EP4, adenosine A2Ar, TGF-β, ACC and NAS. In NK cells and CD8+ T cells, factors inhibiting acetyl-CoA, such as HIF-1α/Ku80 and PDK1 inducers, will inhibit the TCA cycle, OXPHOS and the co-ordination of OXPHOS with glycolysis, in association with a decreased acetylation of COX2. OXPHOS and glycolysis are co-ordinated by a decreased acetylation of Raptor within the mTORC1 complex. The changes in mitochondrial function driven by decreased melatonin, acetyl-CoA, Sirtuins, SOD2, OXPHOS and AMPK will modulate gene expression, partly via levels of ROS production and the associated changes in miRNAs expressed. Altered miRNA patterning will drive variations in the classical immune checkpoint inhibitors, such as the PD-1 suppression by miR-138. Variations in such acetyl-CoA, melatonergic pathway regulation of OXPHOS and glycolysis will occur in the array of immune cells within the tumour microenvironment, with the dynamic intracellular and intercellular interactions of these cells driven by variations in metabolic regulation. Other factors may also ‘backward’ convert melatonin to NAS, including activation of the P2Y1 and mGluR5 receptors, as well as O-demethylation, which are not shown for clarity. The processes highlighted in the figure indicate an important AhR role in the regulation of mitochondrial function. However, the temporal and functional specifics of this await determination in the different cells of the tumour microenvironment, including the interactions of AhR, O-GlcNAcylation, acetyl-CoA and melatonergic pathway. The interactions of AhR effects on mitochondrial function with wider AhR effects on transcription in the different cells of the tumour microenvironment will also be important to determine. Arrows indicate increases/induces; T-arrows indicate suppresses. Abbreviations: AANAT: aralkylamine N-acetyltransferase; ACC: acetyl-CoA carboxylase; acetyl-CoA: acetyl-coenzyme A; AhR: aryl hydrocarbon receptor; AMPK: AMP-activated protein kinase; ASMT: acetylserotonin methyltransferase; CD8+: cluster of differentiation 8; COX2: cyclooxygenase 2; CYP: cytochrome P450; EP4: Prostaglandin E2 receptor 4; FICZ: 6-formylindolo[3,2-b]carbazole; HIF: hypoxia-inducible factor; mTORC1: mammalian target of rapamycin complex 1; NK: natural killer; OXPHOS: oxidative phosphorylation; NAS: N-acetylserotonin; PD-1: programmed cell death-1: PD-L1: programmed cell death ligand 1; PDC: pyruvate dehydrogenase complex; PDK: pyruvate dehydrogenase kinase; SOD: superoxide dismutase; TCA: tricarboxylic acid; TCDD: 2,3,7,8-tetrachlorodibenzo-p-dioxin; TGF: transforming growth factor.