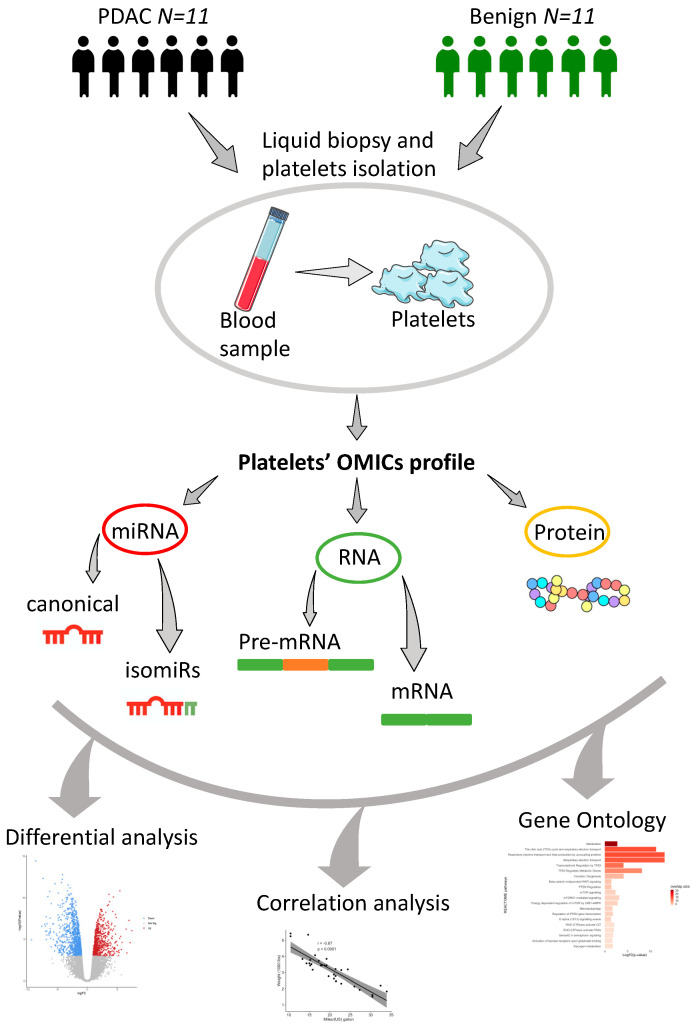
Figure 1.
Workflow of platelets omics profiling. Platelets were isolated from 22 age and sex-matched patients with PDAC and benign diseases. Small-RNAs, RNA transcripts (PDAC samples = 11; benign samples = 11) and proteins (PDAC samples = 8; benign samples = 11) were isolated and sequenced following validated protocols [28,37]. Bioinformatics tools were adopted to quantify canonical as well as isomiRs from smallRNA-seq, mRNA and intron-spanning reads from RNA-seq, and proteins. Downstream analyses were carried out using standalone tools for differential analysis of all data types comparing PDAC versus benign platelets; intra group correlation analysis between miRNAs, mRNAs and proteins of matched PDAC and benign platelets and, lastly, gene ontology mining.