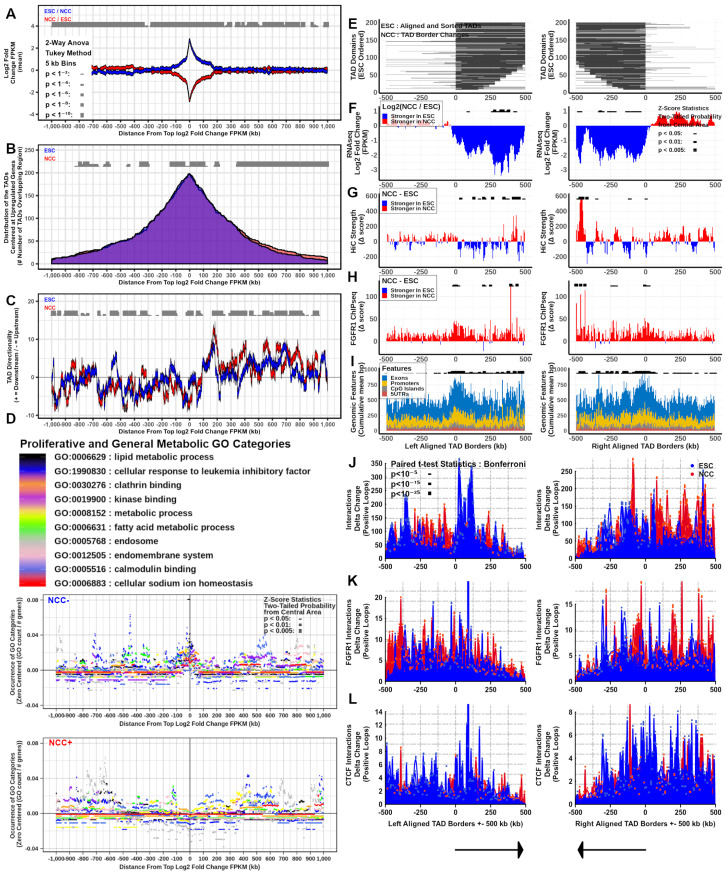
Figure 6.
Downregulated TADs express stage specific genes and are reorganized during ESC to NCC differentiation. At interacting (q < 0.001) upregulated and downregulated anchor-anchor midpoints: (A) Differential gene expression. (B) TAD overlap. (C) Directionality Index. (D) Gene Ontology Category Enrichment. Two-way ANOVA Tukey Method: ESC versus NCC for each location shown, p < 1−2 ; p < 1−4
; p < 1−4 ; p < 1−6
; p < 1−6 ; p < 1−8
; p < 1−8 ; p < 1−10
; p < 1−10 . At TAD borders aligned left and right based on regulated gene directionality (adjusted p < 0.05): (E) TAD reorganization. (F) Differential gene expression. (G) Differential interaction anchor strength. (H) Differential nFGFR1 binding. (I) Differential gene coding and regulator feature enrichment. Z-Score statistics indicate bins which are outside the mean of all bins p < 0.05
. At TAD borders aligned left and right based on regulated gene directionality (adjusted p < 0.05): (E) TAD reorganization. (F) Differential gene expression. (G) Differential interaction anchor strength. (H) Differential nFGFR1 binding. (I) Differential gene coding and regulator feature enrichment. Z-Score statistics indicate bins which are outside the mean of all bins p < 0.05 ; p < 0.01
; p < 0.01 ; p < 0.005
; p < 0.005 . (J) Differential chromatin looping. (K) Differential nFGFR1 looping. (L) Differential CTCF looping. Paired T-Test Bonferroni adjusted p < 10−5
. (J) Differential chromatin looping. (K) Differential nFGFR1 looping. (L) Differential CTCF looping. Paired T-Test Bonferroni adjusted p < 10−5 ; p < 10−15
; p < 10−15 ; p < 10−25
; p < 10−25 .
.